Selection of recombinant MVA by rescue of the essential D4R gene
- PMID: 22152060
- PMCID: PMC3293099
- DOI: 10.1186/1743-422X-8-529
Selection of recombinant MVA by rescue of the essential D4R gene
Abstract
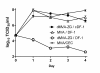
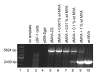
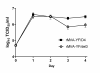
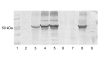
Modified vaccinia virus Ankara (MVA) has become a promising vaccine vector due to its immunogenicity and its proven safety in humans. As a general approach for stringent and rapid selection of recombinant MVA, we assessed marker rescue of the essential viral D4R gene in an engineered deletion mutant that is fully replication defective in wild-type cells. Recombinant, replicating virus was obtained by re-introduction of the deleted viral gene as a dominant selection marker into the deletion mutant.
Figures


Similar articles
-
Expanding the repertoire of Modified Vaccinia Ankara-based vaccine vectors via genetic complementation strategies.PLoS One. 2009;4(5):e5445. doi: 10.1371/journal.pone.0005445. Epub 2009 May 6. PLoS One. 2009. PMID: 19421328 Free PMC article.
-
Spontaneous and Targeted Mutations in the Decapping Enzyme Enhance Replication of Modified Vaccinia Virus Ankara (MVA) in Monkey Cells.J Virol. 2021 Sep 9;95(19):e0110421. doi: 10.1128/JVI.01104-21. Epub 2021 Sep 9. J Virol. 2021. PMID: 34232734 Free PMC article.
-
Defective vaccinia virus as a biologically safe tool for the overproduction of recombinant human secretory proteins.Protein Expr Purif. 1998 Dec;14(3):317-26. doi: 10.1006/prep.1998.0970. Protein Expr Purif. 1998. PMID: 9882565
-
Construction and isolation of recombinant MVA.Methods Mol Biol. 2004;269:77-100. doi: 10.1385/1-59259-789-0:077. Methods Mol Biol. 2004. PMID: 15114009 Review.
-
Use of recombinant vaccinia virus vectors for cell biology.Methods Cell Biol. 1994;43 Pt A:137-59. doi: 10.1016/s0091-679x(08)60602-0. Methods Cell Biol. 1994. PMID: 7823860 Free PMC article. Review.
Cited by
-
MVA vectors expressing conserved influenza proteins protect mice against lethal challenge with H5N1, H9N2 and H7N1 viruses.PLoS One. 2014 Feb 11;9(2):e88340. doi: 10.1371/journal.pone.0088340. eCollection 2014. PLoS One. 2014. PMID: 24523886 Free PMC article.
-
Modified Vaccinia Virus Ankara: History, Value in Basic Research, and Current Perspectives for Vaccine Development.Adv Virus Res. 2017;97:187-243. doi: 10.1016/bs.aivir.2016.07.001. Epub 2016 Aug 1. Adv Virus Res. 2017. PMID: 28057259 Free PMC article. Review.
-
Comparison of Recombinant MVA Selection Methods Based on F13L, D4R and K1L Genes.Viruses. 2022 Mar 4;14(3):528. doi: 10.3390/v14030528. Viruses. 2022. PMID: 35336935 Free PMC article.
-
Identification of β2 microglobulin, the product of B2M gene, as a Host Factor for Vaccinia Virus Infection by Genome-Wide CRISPR genetic screens.PLoS Pathog. 2022 Dec 27;18(12):e1010800. doi: 10.1371/journal.ppat.1010800. eCollection 2022 Dec. PLoS Pathog. 2022. PMID: 36574441 Free PMC article.
References
-
- Mayr A, Hochstein-Mintzel V, Stickl H. Abstammung, Eigenschaften und Verwendung des attenuierten Vaccinia-Stammes MVA. Infection. 1975;3:6–14. doi: 10.1007/BF01641272. - DOI
-
- Mayr A, Stickl H, Muller HK, Danner K, Singer H. The smallpox vaccination strain MVA: marker, genetic structure, experience gained with the parenteral vaccination and behavior in organisms with a debilitated defence mechanism (author's transl) Zentralbl Bakteriol B. 1978;167:375–390. - PubMed
-
- Parrino J, McCurdy LH, Larkin BD, Gordon IJ, Rucker SE, Enama ME, Koup RA, Roederer M, Bailer RT, Moodie Z, Gu L, Yan L, Graham BS. Safety, immunogenicity and efficacy of modified vaccinia Ankara (MVA) against Dryvax challenge in vaccinia-naive and vaccinia-immune individuals. Vaccine. 2007;25:1513–1525. doi: 10.1016/j.vaccine.2006.10.047. - DOI - PMC - PubMed
-
- von Krempelhuber A, Vollmar J, Pokorny R, Rapp P, Wulff N, Petzold B, Handley A, Mateo L, Siersbol H, Kollaritsch H, Chaplin P. A randomized, double-blind, dose-finding Phase II study to evaluate immunogenicity and safety of the third generation smallpox vaccine candidate IMVAMUNE. Vaccine. 2010;28:1209–1216. doi: 10.1016/j.vaccine.2009.11.030. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

