Acyl chain specificity of ceramide synthases is determined within a region of 150 residues in the Tram-Lag-CLN8 (TLC) domain
- PMID: 22144673
- PMCID: PMC3270974
- DOI: 10.1074/jbc.M111.280271
Acyl chain specificity of ceramide synthases is determined within a region of 150 residues in the Tram-Lag-CLN8 (TLC) domain
Abstract
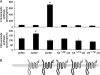
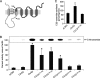
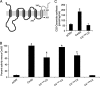
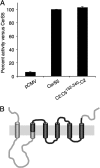
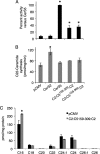
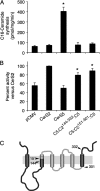
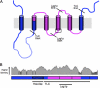
In mammals, ceramides are synthesized by a family of six ceramide synthases (CerS), transmembrane proteins located in the endoplasmic reticulum, where each use fatty acyl-CoAs of defined chain length for ceramide synthesis. Little is known about the molecular features of the CerS that determine acyl-CoA selectivity. We now explore CerS structure-function relationships by constructing chimeric proteins combining sequences from CerS2, which uses C22-CoA for ceramide synthesis, and CerS5, which uses C16-CoA. CerS2 and -5 are 41% identical and 63% similar. Chimeras containing approximately half of CerS5 (from the N terminus) and half of CerS2 (from the C terminus) were catalytically inactive. However, the first 158 residues of CerS5 could be replaced with the equivalent region of CerS2 without affecting specificity of CerS5 toward C16-CoA; likewise, the putative sixth transmembrane domain (at the C terminus) of CerS5 could be replaced with the corresponding sequence of CerS2 without affecting CerS5 specificity. Remarkably, a chimeric CerS5/2 protein containing the first 158 residues and the last 83 residues of CerS2 displayed specificity toward C16-CoA, and a chimeric CerS2/5 protein containing the first 150 residues and the last 79 residues of CerS5 displayed specificity toward C22-CoA, demonstrating that a minimal region of 150 residues is sufficient for retaining CerS specificity.
Figures



Similar articles
-
Eleven residues determine the acyl chain specificity of ceramide synthases.J Biol Chem. 2018 Jun 22;293(25):9912-9921. doi: 10.1074/jbc.RA118.001936. Epub 2018 Apr 9. J Biol Chem. 2018. PMID: 29632068 Free PMC article.
-
A new functional motif in Hox domain-containing ceramide synthases: identification of a novel region flanking the Hox and TLC domains essential for activity.J Biol Chem. 2007 Sep 14;282(37):27366-27373. doi: 10.1074/jbc.M703487200. Epub 2007 Jul 2. J Biol Chem. 2007. PMID: 17609214
-
Characterization of ceramide synthase 2: tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate.J Biol Chem. 2008 Feb 29;283(9):5677-84. doi: 10.1074/jbc.M707386200. Epub 2007 Dec 28. J Biol Chem. 2008. PMID: 18165233
-
Ceramide synthases: roles in cell physiology and signaling.Adv Exp Med Biol. 2010;688:60-71. doi: 10.1007/978-1-4419-6741-1_4. Adv Exp Med Biol. 2010. PMID: 20919646 Review.
-
Synthesis and degradation pathways, functions, and pathology of ceramides and epidermal acylceramides.Prog Lipid Res. 2016 Jul;63:50-69. doi: 10.1016/j.plipres.2016.04.001. Epub 2016 Apr 21. Prog Lipid Res. 2016. PMID: 27107674 Review.
Cited by
-
Interrelations of Sphingolipid and Lysophosphatidate Signaling with Immune System in Ovarian Cancer.Comput Struct Biotechnol J. 2019 Apr 10;17:537-560. doi: 10.1016/j.csbj.2019.04.004. eCollection 2019. Comput Struct Biotechnol J. 2019. PMID: 31049165 Free PMC article.
-
Ceramide-orchestrated signalling in cancer cells.Nat Rev Cancer. 2013 Jan;13(1):51-65. doi: 10.1038/nrc3398. Epub 2012 Dec 13. Nat Rev Cancer. 2013. PMID: 23235911 Review.
-
Glycosphingolipid metabolism and its role in ageing and Parkinson's disease.Glycoconj J. 2022 Feb;39(1):39-53. doi: 10.1007/s10719-021-10023-x. Epub 2021 Nov 10. Glycoconj J. 2022. PMID: 34757540 Free PMC article. Review.
-
Role of Ceramide from Glycosphingolipids and Its Metabolites in Immunological and Inflammatory Responses in Humans.Mediators Inflamm. 2015;2015:120748. doi: 10.1155/2015/120748. Epub 2015 Nov 1. Mediators Inflamm. 2015. PMID: 26609196 Free PMC article. Review.
-
Alternative splicing of ceramide synthase 2 alters levels of specific ceramides and modulates cancer cell proliferation and migration in Luminal B breast cancer subtype.Cell Death Dis. 2021 Feb 10;12(2):171. doi: 10.1038/s41419-021-03436-x. Cell Death Dis. 2021. PMID: 33568634 Free PMC article.
References
-
- Hannun Y. A., Obeid L. M. (2002) The ceramide-centric universe of lipid-mediated cell regulation. Stress encounters of the lipid kind. J. Biol. Chem. 277, 25847–25850 - PubMed
-
- Futerman A. H., Riezman H. (2005) The ins and outs of sphingolipid synthesis. Trends Cell Biol. 15, 312–318 - PubMed
-
- Pewzner-Jung Y., Ben-Dor S., Futerman A. H. (2006) When do Lasses (longevity assurance genes) become CerS (ceramide synthases)? Insights into the regulation of ceramide synthesis. J. Biol. Chem. 281, 25001–25005 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

