Paramutation: just a curiosity or fine tuning of gene expression in the next generation?
- PMID: 22131875
- PMCID: PMC3131737
- DOI: 10.2174/138920211795860099
Paramutation: just a curiosity or fine tuning of gene expression in the next generation?
Abstract
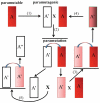
Gene silencing is associated with heritable changes in gene expression which occur without changes in DNA sequence. In eukaryotes these phenomena are common and control important processes, such as development, imprinting, viral and transposon sequence silencing, as well as transgene silencing. Among the epigenetic events, paramutation occurs when a silenced allele (named paramutagenic) is able to silence another allele (paramutable) in trans and this change is heritable. The silenced paramutable allele acquires paramutagenic capacity in the next generations. In the 1950s, Alexander Brink described for the first time the phenomenon of paramutation, occurring in maize at the colored1 (r1) gene, a complex locus (encoding myc-homologous transcription factors) that regulates the anthocyanin biosynthetic pathway. Since then, paramutation and paramutation-like interactions have been discovered in other plants and animals, suggesting that they may underlie important mechanisms for gene expression. The molecular bases of these phenomena are unknown. However in some cases, the event of paramutation has been correlated with changes in DNA methylation, chromatin structure and recently several studies suggest that RNA could play a fundamental role. This last consideration is greatly supported by genetic screening for mutants inhibiting paramutation, which allowed the identification of genes involved in RNA-directed transcriptional silencing, although it is possible that proteins are also required for paramutation.The meaning of paramutation in the life cycle and in evolution remains to be determined even though we might conjecture that this phenomenon could be involved in a fast heritability of favourable epigenetic states across generations in a non-Mendelian way.
Keywords: DNA methylation; Epigenetics; RNA-directed transcriptional silencing.; gene silencing; paramutation; repeated sequences.
Figures

Similar articles
-
Paramutation phenomena in plants.Semin Cell Dev Biol. 2015 Aug;44:2-10. doi: 10.1016/j.semcdb.2015.08.015. Epub 2015 Aug 31. Semin Cell Dev Biol. 2015. PMID: 26335267 Review.
-
Paramutation phenomena in non-vertebrate animals.Semin Cell Dev Biol. 2015 Aug;44:39-46. doi: 10.1016/j.semcdb.2015.08.009. Epub 2015 Aug 28. Semin Cell Dev Biol. 2015. PMID: 26318740 Review.
-
Cis-acting determinants of paramutation.Semin Cell Dev Biol. 2015 Aug;44:22-32. doi: 10.1016/j.semcdb.2015.08.012. Epub 2015 Aug 29. Semin Cell Dev Biol. 2015. PMID: 26321497 Review.
-
Paramutation in maize: RNA mediated trans-generational gene silencing.Curr Opin Genet Dev. 2010 Apr;20(2):156-63. doi: 10.1016/j.gde.2010.01.008. Epub 2010 Feb 12. Curr Opin Genet Dev. 2010. PMID: 20153628 Free PMC article. Review.
-
Transcribed enhancer sequences are required for maize p1 paramutation.Genetics. 2024 Jan 3;226(1):iyad178. doi: 10.1093/genetics/iyad178. Genetics. 2024. PMID: 38169343 Free PMC article.
Cited by
-
Trans-chromosomal methylation.Epigenetics. 2012 Aug;7(8):800-5. doi: 10.4161/epi.20820. Epub 2012 Jun 18. Epigenetics. 2012. PMID: 22705969 Free PMC article. Review.
-
Inheritance of Trans Chromosomal Methylation patterns from Arabidopsis F1 hybrids.Proc Natl Acad Sci U S A. 2014 Feb 4;111(5):2017-22. doi: 10.1073/pnas.1323656111. Epub 2014 Jan 21. Proc Natl Acad Sci U S A. 2014. PMID: 24449910 Free PMC article.
-
Epimutations and mutations, nurturing phenotypic diversity.Genetica. 2022 Aug;150(3-4):171-181. doi: 10.1007/s10709-021-00124-8. Epub 2021 Jun 10. Genetica. 2022. PMID: 34114171 Review.
-
Epigenetic Changes in Hybrids.Plant Physiol. 2015 Aug;168(4):1197-205. doi: 10.1104/pp.15.00231. Epub 2015 May 22. Plant Physiol. 2015. PMID: 26002907 Free PMC article. Review.
-
Anthocyanins in corn: a wealth of genes for human health.Planta. 2014 Nov;240(5):901-11. doi: 10.1007/s00425-014-2131-1. Epub 2014 Aug 9. Planta. 2014. PMID: 25106530 Review.
References
-
- Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. - PubMed
-
- McClintock B. Some parallels between gene control systems in maize and in bacteria. Am. Nat. 1961;95:265–277.
-
- Comfort NC. From controlling elements to transposons: Barbara McClintock and the Nobel Prize. Trends Genet. 2001;17(8):475–478. - PubMed
-
- Russo VEA, Martienssen RA, Riggs AD. Epigenetic Mechanisms of Gene Regulation. Woodbury, USA: Cold Spring Harbor Laboratory Press; 1996.
-
- Martienssen R. Epigenetic phenomena: paramutation and gene silencing in plants. Curr. Biol. 1996;6:810–813. - PubMed
LinkOut - more resources
Full Text Sources
