Automated hydrogen/deuterium exchange electron transfer dissociation high resolution mass spectrometry measured at single-amide resolution
- PMID: 22131230
- PMCID: PMC3796066
- DOI: 10.1007/s13361-011-0298-2
Automated hydrogen/deuterium exchange electron transfer dissociation high resolution mass spectrometry measured at single-amide resolution
Abstract
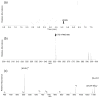
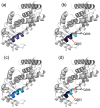
Hydrogen deuterium exchange mass spectrometry (HDX-MS) is a well established method for the measurement of solution-phase deuterium incorporation into proteins, which can provide insight into protein conformational mobility. However, most HDX measurements are constrained to regions of the protein where pepsin proteolysis allows detection at peptide resolution. Recently, single-amide resolution deuterium incorporation has been achieved by limiting gas-phase scrambling in the mass spectrometer. This was accomplished by employing a combination of soft ionization and desolvation conditions coupled with the radical-driven fragmentation technique electron transfer dissociation (ETD). Here, a hybrid LTQ-Orbitrap XL is systematically evaluated for its utility in providing single-amide deuterium incorporation for differential HDX analysis of a nuclear receptor upon binding small molecule ligands. We are able to show that instrumental parameters can be optimized to minimize scrambling and can be incorporated into an established and fully automated HDX platform making differential single-amide HDX possible for bottom-up analysis of complex systems. We have applied this system to determine differential single amide resolution HDX data for the peroxizome proliferator activated receptor bound with two ligands of interest.
© American Society for Mass Spectrometry, 2011
Figures



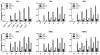
Similar articles
-
Determination of Backbone Amide Hydrogen Exchange Rates of Cytochrome c Using Partially Scrambled Electron Transfer Dissociation Data.J Am Soc Mass Spectrom. 2018 May;29(5):989-1001. doi: 10.1007/s13361-018-1892-3. Epub 2018 Mar 2. J Am Soc Mass Spectrom. 2018. PMID: 29500740
-
Regio-Selective Intramolecular Hydrogen/Deuterium Exchange in Gas-Phase Electron Transfer Dissociation.J Am Soc Mass Spectrom. 2017 May;28(5):971-977. doi: 10.1007/s13361-017-1612-4. Epub 2017 Feb 13. J Am Soc Mass Spectrom. 2017. PMID: 28194737
-
Measuring the hydrogen/deuterium exchange of proteins at high spatial resolution by mass spectrometry: overcoming gas-phase hydrogen/deuterium scrambling.Acc Chem Res. 2014 Oct 21;47(10):3018-27. doi: 10.1021/ar500194w. Epub 2014 Aug 29. Acc Chem Res. 2014. PMID: 25171396 Review.
-
Installation, validation, and application examples of two instrumental setups for gas-phase HDX-MS analysis of peptides and proteins.Methods. 2018 Jul 15;144:113-124. doi: 10.1016/j.ymeth.2018.05.002. Epub 2018 May 18. Methods. 2018. PMID: 29753788
-
Characterizing rapid, activity-linked conformational transitions in proteins via sub-second hydrogen deuterium exchange mass spectrometry.FEBS J. 2013 Nov;280(22):5616-25. doi: 10.1111/febs.12332. Epub 2013 Jun 11. FEBS J. 2013. PMID: 23663649 Review.
Cited by
-
Protein Crystallography in Vaccine Research and Development.Int J Mol Sci. 2015 Jun 9;16(6):13106-40. doi: 10.3390/ijms160613106. Int J Mol Sci. 2015. PMID: 26068237 Free PMC article. Review.
-
A new approach to measuring protein backbone protection with high spatial resolution using H/D exchange and electron capture dissociation.Anal Chem. 2013 Oct 1;85(19):9173-80. doi: 10.1021/ac401868b. Epub 2013 Sep 12. Anal Chem. 2013. PMID: 23978257 Free PMC article.
-
HDX-MS guided drug discovery: small molecules and biopharmaceuticals.Curr Opin Struct Biol. 2014 Oct;28:105-11. doi: 10.1016/j.sbi.2014.08.007. Epub 2014 Aug 30. Curr Opin Struct Biol. 2014. PMID: 25179005 Free PMC article. Review.
-
Glucagon-like peptide-1 receptor ligand interactions: structural cross talk between ligands and the extracellular domain.PLoS One. 2014 Sep 2;9(9):e105683. doi: 10.1371/journal.pone.0105683. eCollection 2014. PLoS One. 2014. PMID: 25180755 Free PMC article.
-
Determination of Equine Cytochrome c Backbone Amide Hydrogen/Deuterium Exchange Rates by Mass Spectrometry Using a Wider Time Window and Isotope Envelope.J Am Soc Mass Spectrom. 2017 Mar;28(3):486-497. doi: 10.1007/s13361-016-1571-1. Epub 2017 Jan 20. J Am Soc Mass Spectrom. 2017. PMID: 28108962
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

