Makorin ring zinc finger protein 1 (MKRN1), a novel poly(A)-binding protein-interacting protein, stimulates translation in nerve cells
- PMID: 22128154
- PMCID: PMC3256852
- DOI: 10.1074/jbc.M111.315291
Makorin ring zinc finger protein 1 (MKRN1), a novel poly(A)-binding protein-interacting protein, stimulates translation in nerve cells
Abstract
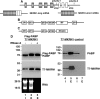
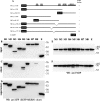
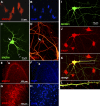
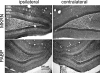
The poly(A)-binding protein (PABP), a key component of different ribonucleoprotein complexes, plays a crucial role in the control of mRNA translation rates, stability, and subcellular targeting. In this study we identify RING zinc finger protein Makorin 1 (MKRN1), a bona fide RNA-binding protein, as a binding partner of PABP that interacts with PABP in an RNA-independent manner. In rat brain, a so far uncharacterized short MKRN1 isoform, MKRN1-short, predominates and is detected in forebrain nerve cells. In neuronal dendrites, MKRN1-short co-localizes with PABP in granule-like structures, which are morphological correlates of sites of mRNA metabolism. Moreover, in primary rat neurons MKRN1-short associates with dendritically localized mRNAs. When tethered to a reporter mRNA, MKRN1-short significantly enhances reporter protein synthesis. Furthermore, after induction of synaptic plasticity via electrical stimulation of the perforant path in vivo, MKRN1-short specifically accumulates in the activated dendritic lamina, the middle molecular layer of the hippocampal dentate gyrus. Collectively, these data indicate that in mammalian neurons MKRN1-short interacts with PABP to locally control the translation of dendritic mRNAs at synapses.
Figures
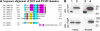


Similar articles
-
The RNA-binding ubiquitin ligase MKRN1 functions in ribosome-associated quality control of poly(A) translation.Genome Biol. 2019 Oct 22;20(1):216. doi: 10.1186/s13059-019-1814-0. Genome Biol. 2019. PMID: 31640799 Free PMC article.
-
Localization of mRNAs at synaptic sites on dendrites.Results Probl Cell Differ. 2001;34:1-26. doi: 10.1007/978-3-540-40025-7_1. Results Probl Cell Differ. 2001. PMID: 11288670 Review. No abstract available.
-
Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation.Neuron. 2001 Apr;30(1):227-40. doi: 10.1016/s0896-6273(01)00275-6. Neuron. 2001. PMID: 11343657
-
Dendritic mRNA targeting and translation.Adv Exp Med Biol. 2012;970:285-305. doi: 10.1007/978-3-7091-0932-8_13. Adv Exp Med Biol. 2012. PMID: 22351061 Review.
-
Cataloguing and Selection of mRNAs Localized to Dendrites in Neurons and Regulated by RNA-Binding Proteins in RNA Granules.Biomolecules. 2020 Jan 22;10(2):167. doi: 10.3390/biom10020167. Biomolecules. 2020. PMID: 31978946 Free PMC article. Review.
Cited by
-
MRKNs: Gene, Functions, and Role in Disease and Infection.Front Oncol. 2022 Apr 8;12:862206. doi: 10.3389/fonc.2022.862206. eCollection 2022. Front Oncol. 2022. PMID: 35463379 Free PMC article. Review.
-
Genome-wide analysis of genes encoding core components of the ubiquitin system during cerebral cortex development.Mol Brain. 2022 Aug 16;15(1):72. doi: 10.1186/s13041-022-00958-z. Mol Brain. 2022. PMID: 35974412 Free PMC article.
-
Crosstalk between ubiquitination and translation in neurodevelopmental disorders.Front Mol Neurosci. 2024 Sep 2;17:1398048. doi: 10.3389/fnmol.2024.1398048. eCollection 2024. Front Mol Neurosci. 2024. PMID: 39286313 Free PMC article. Review.
-
MKRN expression pattern during embryonic and post-embryonic organogenesis in rice (Oryza sativa L. var. Nipponbare).Planta. 2013 Apr;237(4):1083-95. doi: 10.1007/s00425-012-1828-2. Epub 2012 Dec 23. Planta. 2013. PMID: 23262670
-
Inhibition of Poly(A)-binding protein with a synthetic RNA mimic reduces pain sensitization in mice.Nat Commun. 2018 Jan 2;9(1):10. doi: 10.1038/s41467-017-02449-5. Nat Commun. 2018. PMID: 29295980 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

