Molecular basis of dihydrouridine formation on tRNA
- PMID: 22123979
- PMCID: PMC3241823
- DOI: 10.1073/pnas.1112352108
Molecular basis of dihydrouridine formation on tRNA
Abstract
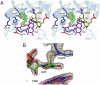
Dihydrouridine (D) is a highly conserved modified base found in tRNAs from all domains of life. Dihydrouridine synthase (Dus) catalyzes the D formation of tRNA through reduction of uracil base with flavin mononucleotide (FMN) as a cofactor. Here, we report the crystal structures of Thermus thermophilus Dus (TthDus), which is responsible for D formation at positions 20 and 20a, in complex with tRNA and with a short fragment of tRNA (D-loop). Dus interacts extensively with the D-arm and recognizes the elbow region composed of the kissing loop interaction between T- and D-loops in tRNA, pulling U20 into the catalytic center for reduction. Although distortion of the D-loop structure was observed upon binding of Dus to tRNA, the canonical D-loop/T-loop interaction was maintained. These results were consistent with the observation that Dus preferentially recognizes modified rather than unmodified tRNAs, indicating that Dus introduces D20 by monitoring the complete L-shaped structure of tRNAs. In the active site, U20 is stacked on the isoalloxazine ring of FMN, and C5 of the U20 uracil ring is covalently cross linked to the thiol group of Cys93, implying a catalytic mechanism of D20 formation. In addition, the involvement of a cofactor molecule in uracil ring recognition was proposed. Based on a series of mutation analyses, we propose a molecular basis of tRNA recognition and D formation catalyzed by Dus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
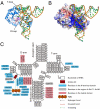
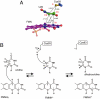
 ). (C) Overall interaction scheme among TthDus, FMN, and tRNA. Hydrogen bonds, π-stacking interaction, and covalent bond between TthDus and tRNA are shown as black, green, and red arrows, respectively.
). (C) Overall interaction scheme among TthDus, FMN, and tRNA. Hydrogen bonds, π-stacking interaction, and covalent bond between TthDus and tRNA are shown as black, green, and red arrows, respectively.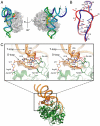
Similar articles
-
In vitro dihydrouridine formation by tRNA dihydrouridine synthase from Thermus thermophilus, an extreme-thermophilic eubacterium.J Biochem. 2015 Dec;158(6):513-21. doi: 10.1093/jb/mvv066. Epub 2015 Jun 24. J Biochem. 2015. PMID: 26112661
-
Crystallization and preliminary X-ray crystallographic analysis of dihydrouridine synthase from Thermus thermophilus and its complex with tRNA.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011 Jun 1;67(Pt 6):685-8. doi: 10.1107/S1744309111012486. Epub 2011 May 25. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011. PMID: 21636912 Free PMC article.
-
Unveiling structural and functional divergences of bacterial tRNA dihydrouridine synthases: perspectives on the evolution scenario.Nucleic Acids Res. 2018 Feb 16;46(3):1386-1394. doi: 10.1093/nar/gkx1294. Nucleic Acids Res. 2018. PMID: 29294097 Free PMC article.
-
The Dihydrouridine landscape from tRNA to mRNA: a perspective on synthesis, structural impact and function.RNA Biol. 2022 Jan;19(1):735-750. doi: 10.1080/15476286.2022.2078094. RNA Biol. 2022. PMID: 35638108 Free PMC article. Review.
-
The m1A(58) modification in eubacterial tRNA: An overview of tRNA recognition and mechanism of catalysis by TrmI.Biophys Chem. 2016 Mar;210:27-34. doi: 10.1016/j.bpc.2015.06.012. Epub 2015 Jul 16. Biophys Chem. 2016. PMID: 26189113 Review.
Cited by
-
Regulation of the epigenome through RNA modifications.Chromosoma. 2023 Sep;132(3):231-246. doi: 10.1007/s00412-023-00794-7. Epub 2023 May 4. Chromosoma. 2023. PMID: 37138119 Free PMC article. Review.
-
Molecular evolution of dihydrouridine synthases.BMC Bioinformatics. 2012 Jun 28;13:153. doi: 10.1186/1471-2105-13-153. BMC Bioinformatics. 2012. PMID: 22741570 Free PMC article.
-
Identification of D Modification Sites Using a Random Forest Model Based on Nucleotide Chemical Properties.Int J Mol Sci. 2022 Mar 11;23(6):3044. doi: 10.3390/ijms23063044. Int J Mol Sci. 2022. PMID: 35328461 Free PMC article.
-
RNA modifications and the link to human disease.Methods Enzymol. 2019;626:133-146. doi: 10.1016/bs.mie.2019.08.003. Epub 2019 Sep 3. Methods Enzymol. 2019. PMID: 31606073 Free PMC article. Review.
-
Transfer RNA Modification Enzymes from Thermophiles and Their Modified Nucleosides in tRNA.Microorganisms. 2018 Oct 20;6(4):110. doi: 10.3390/microorganisms6040110. Microorganisms. 2018. PMID: 30347855 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

