An exogenous chloroplast genome for complex sequence manipulation in algae
- PMID: 22116061
- PMCID: PMC3315318
- DOI: 10.1093/nar/gkr1008
An exogenous chloroplast genome for complex sequence manipulation in algae
Abstract
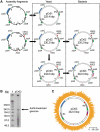
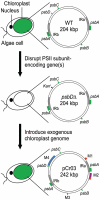
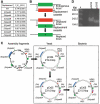
We demonstrate a system for cloning and modifying the chloroplast genome from the green alga, Chlamydomonas reinhardtii. Through extensive use of sequence stabilization strategies, the ex vivo genome is assembled in yeast from a collection of overlapping fragments. The assembled genome is then moved into bacteria for large-scale preparations and transformed into C. reinhardtii cells. This system also allows for the generation of simultaneous, systematic and complex genetic modifications at multiple loci in vivo. We use this system to substitute genes encoding core subunits of the photosynthetic apparatus with orthologs from a related alga, Scenedesmus obliquus. Once transformed into algae, the substituted genome recombines with the endogenous genome, resulting in a hybrid plastome comprising modifications in disparate loci. The in vivo function of the genomes described herein demonstrates that simultaneous engineering of multiple sites within the chloroplast genome is now possible. This work represents the first steps toward a novel approach for creating genetic diversity in any or all regions of a chloroplast genome.
Figures
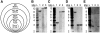
Similar articles
-
Refactoring the Six-Gene Photosystem II Core in the Chloroplast of the Green Algae Chlamydomonas reinhardtii.ACS Synth Biol. 2016 Jul 15;5(7):589-96. doi: 10.1021/acssynbio.5b00076. Epub 2015 Aug 7. ACS Synth Biol. 2016. PMID: 26214707
-
Production of Recombinant Proteins in the Chloroplast of the Green Alga Chlamydomonas reinhardtii.Methods Mol Biol. 2016;1385:69-85. doi: 10.1007/978-1-4939-3289-4_5. Methods Mol Biol. 2016. PMID: 26614282
-
A simple, low-cost method for chloroplast transformation of the green alga Chlamydomonas reinhardtii.Methods Mol Biol. 2014;1132:401-11. doi: 10.1007/978-1-62703-995-6_27. Methods Mol Biol. 2014. PMID: 24599870
-
Chlamydomonas reinhardtii as the photosynthetic yeast.Annu Rev Genet. 1995;29:209-30. doi: 10.1146/annurev.ge.29.120195.001233. Annu Rev Genet. 1995. PMID: 8825474 Review.
-
Tools and techniques for chloroplast transformation of Chlamydomonas.Adv Exp Med Biol. 2007;616:34-45. doi: 10.1007/978-0-387-75532-8_4. Adv Exp Med Biol. 2007. PMID: 18161489 Review.
Cited by
-
Recent Advances and Current Challenges in Synthetic Biology of the Plastid Genetic System and Metabolism.Plant Physiol. 2019 Mar;179(3):794-802. doi: 10.1104/pp.18.00767. Epub 2018 Sep 4. Plant Physiol. 2019. PMID: 30181342 Free PMC article. Review.
-
Plant Biosystems Design Research Roadmap 1.0.Biodes Res. 2020 Dec 5;2020:8051764. doi: 10.34133/2020/8051764. eCollection 2020. Biodes Res. 2020. PMID: 37849899 Free PMC article. Review.
-
Green biologics: The algal chloroplast as a platform for making biopharmaceuticals.Bioengineered. 2018 Jan 1;9(1):48-54. doi: 10.1080/21655979.2017.1377867. Epub 2017 Sep 29. Bioengineered. 2018. PMID: 28892417 Free PMC article. Review.
-
Cloning of Thalassiosira pseudonana's Mitochondrial Genome in Saccharomyces cerevisiae and Escherichia coli.Biology (Basel). 2020 Oct 26;9(11):358. doi: 10.3390/biology9110358. Biology (Basel). 2020. PMID: 33114477 Free PMC article.
-
Genomics of Volvocine Algae.Adv Bot Res. 2012;64:185-243. doi: 10.1016/B978-0-12-391499-6.00006-2. Adv Bot Res. 2012. PMID: 25883411 Free PMC article.
References
-
- Carr PA, Church GM. Genome engineering. Nat. Biotechnol. 2009;27:1151–1162. - PubMed
-
- Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, et al. Complete Chemical Synthesis, Assembly, and Cloning of a Mycoplasma genitalium Genome. Science. 2008;319:1215–1220. - PubMed
-
- Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang R-Y, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, et al. Creation of a Bacterial Cell Controlled by a Chemically Synthesized Genome. Science. 2010;329:52–56. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

