Hes6 is required for the neurogenic activity of neurogenin and NeuroD
- PMID: 22114720
- PMCID: PMC3218063
- DOI: 10.1371/journal.pone.0027880
Hes6 is required for the neurogenic activity of neurogenin and NeuroD
Abstract
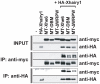
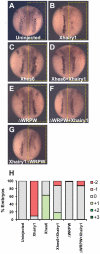
In the embryonic neural plate, a subset of precursor cells with neurogenic potential differentiates into neurons. This process of primary neurogenesis requires both the specification of cells for neural differentiation, regulated by Notch signaling, and the activity of neurogenic transcription factors such as neurogenin and NeuroD which drive the program of neural gene expression. Here we study the role of Hes6, a member of the hairy enhancer of split family of transcription factors, in primary neurogenesis in Xenopus embryos. Hes6 is an atypical Hes gene in that it is not regulated by Notch signaling and promotes neural differentiation in mouse cell culture models. We show that depletion of Xenopus Hes6 (Xhes6) by morpholino antisense oligonucleotides results in a failure of neural differentiation, a phenotype rescued by both wild type Xhes6 and a Xhes6 mutant unable to bind DNA. However, an Xhes6 mutant that lacks the ability to bind Groucho/TLE transcriptional co-regulators is only partly able to rescue the phenotype. Further analysis reveals that Xhes6 is essential for the induction of neurons by both neurogenin and NeuroD, acting via at least two distinct mechanisms, the inhibition of antineurogenic Xhairy proteins and by interaction with Groucho/TLE family proteins. We conclude Xhes6 is essential for neurogenesis in vivo, acting via multiple mechanisms to relieve inhibition of proneural transcription factor activity within the neural plate.
Conflict of interest statement
Figures

Similar articles
-
The Groucho/Transducin-like enhancer of split protein family in animal development.IUBMB Life. 2015 Jul;67(7):472-81. doi: 10.1002/iub.1395. Epub 2015 Jul 14. IUBMB Life. 2015. PMID: 26172616 Free PMC article. Review.
-
Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers.EMBO J. 2007 Dec 12;26(24):5093-108. doi: 10.1038/sj.emboj.7601923. Epub 2007 Nov 15. EMBO J. 2007. PMID: 18007592 Free PMC article.
-
Hes6 is required for MyoD induction during gastrulation.Dev Biol. 2007 Dec 1;312(1):61-76. doi: 10.1016/j.ydbio.2007.09.011. Epub 2007 Sep 16. Dev Biol. 2007. PMID: 17950722
-
Hes6 acts in a positive feedback loop with the neurogenins to promote neuronal differentiation.Development. 2000 Oct;127(19):4203-16. doi: 10.1242/dev.127.19.4203. Development. 2000. PMID: 10976052
-
Making neurons, made easy: The use of Neurogenin-2 in neuronal differentiation.Stem Cell Reports. 2022 Jan 11;17(1):14-34. doi: 10.1016/j.stemcr.2021.11.015. Epub 2021 Dec 30. Stem Cell Reports. 2022. PMID: 34971564 Free PMC article. Review.
Cited by
-
HES6 drives a critical AR transcriptional programme to induce castration-resistant prostate cancer through activation of an E2F1-mediated cell cycle network.EMBO Mol Med. 2014 May;6(5):651-61. doi: 10.1002/emmm.201303581. EMBO Mol Med. 2014. PMID: 24737870 Free PMC article.
-
The emerging roles of Notch signaling in leukemia and stem cells.Biomark Res. 2013 Jul 18;1(1):23. doi: 10.1186/2050-7771-1-23. Biomark Res. 2013. PMID: 24252593 Free PMC article.
-
Notch Signaling in Pancreatic Development.Int J Mol Sci. 2015 Dec 30;17(1):48. doi: 10.3390/ijms17010048. Int J Mol Sci. 2015. PMID: 26729103 Free PMC article. Review.
-
N-terminal phosphorylation of xHes1 controls inhibition of primary neurogenesis in Xenopus.Biochem Biophys Res Commun. 2019 Feb 5;509(2):557-563. doi: 10.1016/j.bbrc.2018.12.135. Epub 2018 Dec 29. Biochem Biophys Res Commun. 2019. PMID: 30600182 Free PMC article.
-
The Groucho/Transducin-like enhancer of split protein family in animal development.IUBMB Life. 2015 Jul;67(7):472-81. doi: 10.1002/iub.1395. Epub 2015 Jul 14. IUBMB Life. 2015. PMID: 26172616 Free PMC article. Review.
References
-
- Hartenstein V. Early neurogenesis in Xenopus: the spatio-temporal pattern of proliferation and cell lineages in the embryonic spinal cord. Neuron. 1989;3:399–411. - PubMed
-
- Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 1995;375:761–766. - PubMed
-
- Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. - PubMed
-
- Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, et al. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. - PubMed
-
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

