VDR/RXR and TCF4/β-catenin cistromes in colonic cells of colorectal tumor origin: impact on c-FOS and c-MYC gene expression
- PMID: 22108803
- PMCID: PMC3248320
- DOI: 10.1210/me.2011-1109
VDR/RXR and TCF4/β-catenin cistromes in colonic cells of colorectal tumor origin: impact on c-FOS and c-MYC gene expression
Abstract
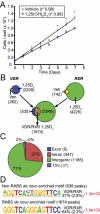
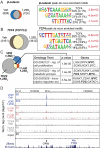
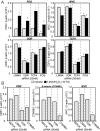
Many of the transcriptional and growth regulating activities of 1α,25-dihydroxyvitamin D(3) [1,25-(OH)(2)D(3)] in the intestine and colon are recapitulated in the human colorectal cancer cell LS180. We therefore used this line together with chromatin immunoprecipitation-seq and gene expression analyses to identify the vitamin D receptor (VDR)/retinoid X receptor (RXR) and transcription factor 7-like 2 (TCF7L2/TCF4)/β-catenin cistromes and the genes that they regulate. VDR and RXR colocalized to predominantly promoter distal, vitamin D response element-containing sites in a largely ligand-dependent manner. These regulatory sites control the expression of both known as well as novel 1,25-(OH)(2)D(3) target genes. TCF4 and β-catenin cistromes partially overlapped, contained TCF/lymphoid enhancer-binding factor consensus elements, and were only modestly influenced by 1,25-(OH)(2)D(3). However, the two heterodimer complexes colocalized at sites near a limited set of genes that included c-FOS and c-MYC; the expression of both genes was modulated by 1,25-(OH)(2)D(3). At the c-FOS gene, both VDR/RXR and TCF4/β-catenin bound to a single distal enhancer located 24 kb upstream of the transcriptional start site. At the c-MYC locus, however, binding was noted at a cluster of sites between -139 and -165 kb and at a site located -335 kb upstream. Examined as isolated enhancer fragments, these regions exhibited basal and 1,25-(OH)(2)D(3)-inducible activities that were interlinked to both VDR and β-catenin activation. These data reveal additional complexity in the regulation of target genes by 1,25-(OH)(2)D(3) and support a direct action of both VDR and the TCF4/β-catenin regulatory complex at c-FOS and c-MYC.
Figures



Similar articles
-
A dynamic exchange of TCF3 and TCF4 transcription factors controls MYC expression in colorectal cancer cells.Cell Cycle. 2015;14(3):323-32. doi: 10.4161/15384101.2014.980643. Cell Cycle. 2015. PMID: 25659031 Free PMC article.
-
Multifunctional enhancers regulate mouse and human vitamin D receptor gene transcription.Mol Endocrinol. 2010 Jan;24(1):128-47. doi: 10.1210/me.2009-0140. Epub 2009 Nov 6. Mol Endocrinol. 2010. PMID: 19897601 Free PMC article.
-
Control of TCF-4 expression by VDR and vitamin D in the mouse mammary gland and colorectal cancer cell lines.PLoS One. 2009 Nov 17;4(11):e7872. doi: 10.1371/journal.pone.0007872. PLoS One. 2009. PMID: 19924301 Free PMC article.
-
New understanding of the molecular mechanism of receptor-mediated genomic actions of the vitamin D hormone.Bone. 1995 Aug;17(2 Suppl):33S-38S. doi: 10.1016/8756-3282(95)00205-r. Bone. 1995. PMID: 8579895 Review.
-
The vitamin D hormone and its nuclear receptor: molecular actions and disease states.J Endocrinol. 1997 Sep;154 Suppl:S57-73. J Endocrinol. 1997. PMID: 9379138 Review.
Cited by
-
Xenobiotic-sensing nuclear receptors involved in drug metabolism: a structural perspective.Drug Metab Rev. 2013 Feb;45(1):79-100. doi: 10.3109/03602532.2012.740049. Epub 2012 Dec 5. Drug Metab Rev. 2013. PMID: 23210723 Free PMC article. Review.
-
Calcipotriol Targets LRP6 to Inhibit Wnt Signaling in Pancreatic Cancer.Mol Cancer Res. 2015 Nov;13(11):1509-19. doi: 10.1158/1541-7786.MCR-15-0204. Epub 2015 Jul 29. Mol Cancer Res. 2015. PMID: 26224368 Free PMC article.
-
ReMo-SNPs: a new software tool for identification of polymorphisms in regions and motifs genome-wide.Genet Res (Camb). 2015 Apr 17;97:e8. doi: 10.1017/S0016672315000051. Genet Res (Camb). 2015. PMID: 25882789 Free PMC article.
-
Vitamin D and Its Target Genes.Nutrients. 2022 Mar 24;14(7):1354. doi: 10.3390/nu14071354. Nutrients. 2022. PMID: 35405966 Free PMC article. Review.
-
Ex vivo culture of primary human colonic tissue for studying transcriptional responses to 1α,25(OH)2 and 25(OH) vitamin D.Physiol Genomics. 2014 Apr 15;46(8):302-8. doi: 10.1152/physiolgenomics.00194.2013. Epub 2014 Feb 18. Physiol Genomics. 2014. PMID: 24550213 Free PMC article.
References
-
- Plum LA, DeLuca HF. 2010. Vitamin D, disease and therapeutic opportunities. Nat Rev Drug Discov 9:941–955 - PubMed
-
- Bikle DD. 2010. Vitamin D and the skin. J Bone Miner Metab 28:117–130 - PubMed
-
- Deeb KK, Trump DL, Johnson CS. 2007. Vitamin D signalling pathways in cancer: potential for anticancer therapeutics. Nat Rev Cancer 7:684–700 - PubMed
-
- Welsh J. 2004. Vitamin D and breast cancer: insights from animal models. Am J Clin Nutr 80:1721S–1724S - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

