Arabidopsis CSP41 proteins form multimeric complexes that bind and stabilize distinct plastid transcripts
- PMID: 22090436
- PMCID: PMC3276088
- DOI: 10.1093/jxb/err347
Arabidopsis CSP41 proteins form multimeric complexes that bind and stabilize distinct plastid transcripts
Abstract
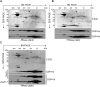
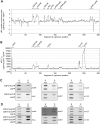
The spinach CSP41 protein has been shown to bind and cleave chloroplast RNA in vitro. Arabidopsis thaliana, like other photosynthetic eukaryotes, encodes two copies of this protein. Several functions have been described for CSP41 proteins in Arabidopsis, including roles in chloroplast rRNA metabolism and transcription. CSP41a and CSP41b interact physically, but it is not clear whether they have distinct functions. It is shown here that CSP41b, but not CSP41a, is an essential and major component of a specific subset of RNA-binding complexes that form in the dark and disassemble in the light. RNA immunoprecipitation and hybridization to gene chips (RIP-chip) experiments indicated that CSP41 complexes can contain chloroplast mRNAs coding for photosynthetic proteins and rRNAs (16S and 23S), but no tRNAs or mRNAs for ribosomal proteins. Leaves of plants lacking CSP41b showed decreased steady-state levels of CSP41 target RNAs, as well as decreased plastid transcription and translation rates. Representative target RNAs were less stable when incubated with broken chloroplasts devoid of CSP41 complexes, indicating that CSP41 proteins can stabilize target RNAs. Therefore, it is proposed that (i) CSP41 complexes may serve to stabilize non-translated target mRNAs and precursor rRNAs during the night when the translational machinery is less active in a manner responsive to the redox state of the chloroplast, and (ii) that the defects in translation and transcription in CSP41 protein-less mutants are secondary effects of the decreased transcript stability.
Figures

Similar articles
-
The conserved endoribonuclease YbeY is required for chloroplast ribosomal RNA processing in Arabidopsis.Plant Physiol. 2015 May;168(1):205-21. doi: 10.1104/pp.114.255000. Epub 2015 Mar 25. Plant Physiol. 2015. PMID: 25810095 Free PMC article.
-
The RNA-binding proteins CSP41a and CSP41b may regulate transcription and translation of chloroplast-encoded RNAs in Arabidopsis.Plant Mol Biol. 2009 Mar;69(5):541-52. doi: 10.1007/s11103-008-9436-z. Epub 2008 Dec 9. Plant Mol Biol. 2009. PMID: 19067181
-
Arabidopsis thaliana mutants reveal a role for CSP41a and CSP41b, two ribosome-associated endonucleases, in chloroplast ribosomal RNA metabolism.Plant Mol Biol. 2008 Jul;67(4):389-401. doi: 10.1007/s11103-008-9328-2. Epub 2008 Apr 9. Plant Mol Biol. 2008. PMID: 18398686
-
The cutting crew - ribonucleases are key players in the control of plastid gene expression.J Exp Bot. 2012 Feb;63(4):1663-73. doi: 10.1093/jxb/err401. Epub 2011 Dec 3. J Exp Bot. 2012. PMID: 22140236 Review.
-
Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover.Prog Nucleic Acid Res Mol Biol. 2004;78:305-37. doi: 10.1016/S0079-6603(04)78008-3. Prog Nucleic Acid Res Mol Biol. 2004. PMID: 15210334 Review.
Cited by
-
ALB3 Insertase Mediates Cytochrome b6 Co-translational Import into the Thylakoid Membrane.Sci Rep. 2016 Oct 4;6:34557. doi: 10.1038/srep34557. Sci Rep. 2016. PMID: 27698412 Free PMC article.
-
The conserved endoribonuclease YbeY is required for chloroplast ribosomal RNA processing in Arabidopsis.Plant Physiol. 2015 May;168(1):205-21. doi: 10.1104/pp.114.255000. Epub 2015 Mar 25. Plant Physiol. 2015. PMID: 25810095 Free PMC article.
-
Organellar and Secretory Ribonucleases: Major Players in Plant RNA Homeostasis.Plant Physiol. 2020 Aug;183(4):1438-1452. doi: 10.1104/pp.20.00076. Epub 2020 Jun 8. Plant Physiol. 2020. PMID: 32513833 Free PMC article.
-
Chloroplast gene expression: Recent advances and perspectives.Plant Commun. 2023 Sep 11;4(5):100611. doi: 10.1016/j.xplc.2023.100611. Epub 2023 May 4. Plant Commun. 2023. PMID: 37147800 Free PMC article. Review.
-
Dawn regulates guard cell proteins in Arabidopsis thaliana that function in ATP production from fatty acid beta-oxidation.Plant Mol Biol. 2018 Dec;98(6):525-543. doi: 10.1007/s11103-018-0794-x. Epub 2018 Nov 3. Plant Mol Biol. 2018. PMID: 30392160
References
-
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. - PubMed
-
- Baker ME, Grundy WN, Elkan CP. Spinach CSP41, an mRNA-binding protein and ribonuclease, is homologous to nucleotide-sugar epimerases and hydroxysteroid dehydrogenases. Biochemical and Biophysical Research Communications. 1998;248:250–254. - PubMed
-
- Barkan A, Goldschmidt-Clermont M. Participation of nuclear genes in chloroplast gene expression. Biochimie. 2000;82:559–572. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

