Aberrant firing of replication origins potentially explains intragenic nonrecurrent rearrangements within genes, including the human DMD gene
- PMID: 22090376
- PMCID: PMC3246204
- DOI: 10.1101/gr.123463.111
Aberrant firing of replication origins potentially explains intragenic nonrecurrent rearrangements within genes, including the human DMD gene
Abstract
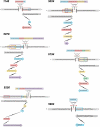
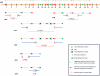
Non-allelic homologous recombination (NAHR), non-homologous end joining (NHEJ), and microhomology-mediated replication-dependent recombination (MMRDR) have all been put forward as mechanisms to explain DNA rearrangements associated with genomic disorders. However, many nonrecurrent rearrangements in humans remain unexplained. To further investigate the mutation mechanisms of these copy number variations (CNVs), we performed breakpoint mapping analysis for 62 clinical cases with intragenic deletions in the human DMD gene (50 cases) and other known disease-causing genes (one PCCB, one IVD, one DBT, three PAH, one STK11, one HEXB, three DBT, one HRPT1, and one EMD cases). While repetitive elements were found in only four individual cases, three involving DMD and one HEXB gene, microhomologies (2-10 bp) were observed at breakpoint junctions in 56% and insertions ranging from 1 to 48 bp were seen in 16 of the total 62 cases. Among these insertions, we observed evidence for tandem repetitions of short segments (5-20 bp) of reference sequence proximal to the breakpoints in six individual DMD cases (six repeats in one, four repeats in three, two repeats in one, and one repeat in one case), strongly indicating attempts by the replication machinery to surpass the stalled replication fork. We provide evidence of a novel template slippage event during replication rescue. With a deeper insight into the complex process of replication and its rescue during origin failure, brought forward by recent studies, we propose a hypothesis based on aberrant firing of replication origins to explain intragenic nonrecurrent rearrangements within genes, including the DMD gene.
Figures


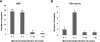
Similar articles
-
Complex genomic rearrangements in the dystrophin gene due to replication-based mechanisms.Mol Genet Genomic Med. 2014 Nov;2(6):539-47. doi: 10.1002/mgg3.108. Epub 2014 Sep 15. Mol Genet Genomic Med. 2014. PMID: 25614876 Free PMC article.
-
Non Random Distribution of DMD Deletion Breakpoints and Implication of Double Strand Breaks Repair and Replication Error Repair Mechanisms.J Neuromuscul Dis. 2016 May 27;3(2):227-245. doi: 10.3233/JND-150134. J Neuromuscul Dis. 2016. PMID: 27854212
-
A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders.Cell. 2007 Dec 28;131(7):1235-47. doi: 10.1016/j.cell.2007.11.037. Cell. 2007. PMID: 18160035
-
Genomic rearrangements in inherited disease and cancer.Semin Cancer Biol. 2010 Aug;20(4):222-33. doi: 10.1016/j.semcancer.2010.05.007. Epub 2010 Jun 9. Semin Cancer Biol. 2010. PMID: 20541013 Review.
-
Origins and breakpoint analyses of copy number variations: up close and personal.Cytogenet Genome Res. 2011;135(3-4):271-6. doi: 10.1159/000330267. Epub 2011 Aug 12. Cytogenet Genome Res. 2011. PMID: 21846967 Review.
Cited by
-
fhl2b mediates extraocular muscle protection in zebrafish models of muscular dystrophies and its ectopic expression ameliorates affected body muscles.Nat Commun. 2024 Mar 2;15(1):1950. doi: 10.1038/s41467-024-46187-x. Nat Commun. 2024. PMID: 38431640 Free PMC article.
-
Genomic and biochemical analysis of repeatedly observed variants in DBT in individuals with maple syrup urine disease of Central American ancestry.Am J Med Genet A. 2022 Sep;188(9):2738-2749. doi: 10.1002/ajmg.a.62893. Epub 2022 Jul 7. Am J Med Genet A. 2022. PMID: 35799415 Free PMC article.
-
Replicative mechanisms of CNV formation preferentially occur as intrachromosomal events: evidence from Potocki-Lupski duplication syndrome.Hum Mol Genet. 2013 Feb 15;22(4):749-56. doi: 10.1093/hmg/dds482. Epub 2012 Nov 16. Hum Mol Genet. 2013. PMID: 23161748 Free PMC article.
-
Duchenne muscular dystrophy trajectory in R-DMDdel52 preclinical rat model identifies COMP as biomarker of fibrosis.Acta Neuropathol Commun. 2022 Apr 25;10(1):60. doi: 10.1186/s40478-022-01355-2. Acta Neuropathol Commun. 2022. PMID: 35468843 Free PMC article.
-
Decoding NF1 Intragenic Copy-Number Variations.Am J Hum Genet. 2015 Aug 6;97(2):238-49. doi: 10.1016/j.ajhg.2015.06.002. Epub 2015 Jul 16. Am J Hum Genet. 2015. PMID: 26189818 Free PMC article.
References
-
- Anglana M, Apiou F, Bensimon A, Debatisse M 2003. Dynamics of DNA replication in mammalian somatic cells: Nucleotide pool modulates origin choice and interorigin spacing. Cell 114: 385–394 - PubMed
-
- Berezney R, Dubey DD, Huberman JA 2000. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108: 471–484 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
