Duck Hepatitis A virus possesses a distinct type IV internal ribosome entry site element of picornavirus
- PMID: 22090106
- PMCID: PMC3255860
- DOI: 10.1128/JVI.00306-11
Duck Hepatitis A virus possesses a distinct type IV internal ribosome entry site element of picornavirus
Abstract
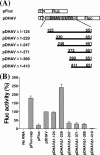
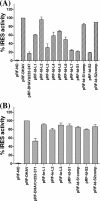
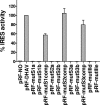
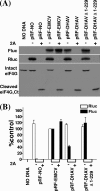
Sequence analysis of duck hepatitis virus type 1 (DHV-1) led to its classification as the only member of a new genus, Avihepatovirus, of the family Picornaviridae, and so was renamed duck hepatitis A virus (DHAV). The 5' untranslated region (5' UTR) plays an important role in translation initiation and RNA synthesis of the picornavirus. Here, we provide evidence that the 651-nucleotide (nt)-long 5' UTR of DHAV genome contains an internal ribosome entry site (IRES) element that functions efficiently in vitro and within BHK cells. Comparative sequence analysis showed that the 3' part of the DHAV 5' UTR is similar to the porcine teschovirus 1 (PTV-1) IRES in sequence and predicted secondary structure. Further mutational analyses of the predicted domain IIId, domain IIIe, and pseudoknot structure at the 3' end of the DHAV IRES support our predicted secondary structure. However, unlike the case for the PTV-1 IRES element, analysis of various deletion mutants demonstrated that the optimally functional DHAV IRES element with a size of approximately 420 nt is larger than that of PTV-1 and contains other peripheral domains (Id and Ie) that do not exist within the type IV IRES elements. The domain Ie, however, could be removed without significant loss of activity. Surprisingly, like the hepatitis A virus (HAV) IRES element, the activity of DHAV IRES could be eliminated by expression of enterovirus 2A protease. These findings indicate that the DHAV IRES shares common features with type IV picornavirus IRES elements, whereas it exhibits significant differences from type IV IRESs. Therefore, we propose that DHAV possesses a distinct type IV IRES element of picornavirus.
Figures





Similar articles
-
The picornavirus avian encephalomyelitis virus possesses a hepatitis C virus-like internal ribosome entry site element.J Virol. 2008 Feb;82(4):1993-2003. doi: 10.1128/JVI.01957-07. Epub 2007 Dec 12. J Virol. 2008. PMID: 18077729 Free PMC article.
-
The duck hepatitis virus 5'-UTR possesses HCV-like IRES activity that is independent of eIF4F complex and modulated by downstream coding sequences.Virol J. 2011 Mar 31;8:147. doi: 10.1186/1743-422X-8-147. Virol J. 2011. PMID: 21450110 Free PMC article.
-
A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: evidence for modular exchange of functional noncoding RNA elements by recombination.J Virol. 2007 Jun;81(11):5850-63. doi: 10.1128/JVI.02403-06. Epub 2007 Mar 28. J Virol. 2007. PMID: 17392358 Free PMC article.
-
Divergent picornavirus IRES elements.Virus Res. 2009 Feb;139(2):183-92. doi: 10.1016/j.virusres.2008.07.001. Epub 2008 Aug 20. Virus Res. 2009. PMID: 18675861 Review.
-
Picornavirus IRES: structure function relationship.Curr Pharm Des. 2004;10(30):3757-67. doi: 10.2174/1381612043382657. Curr Pharm Des. 2004. PMID: 15579069 Review.
Cited by
-
Specific DNAzymes cleave the 300-618 nt of 5'UTR to inhibit DHAV-1 translation and replication.Front Microbiol. 2022 Dec 12;13:1064612. doi: 10.3389/fmicb.2022.1064612. eCollection 2022. Front Microbiol. 2022. PMID: 36578574 Free PMC article.
-
Construction and characterization of an improved DNA-launched infectious clone of duck hepatitis a virus type 1.Virol J. 2017 Nov 3;14(1):212. doi: 10.1186/s12985-017-0883-5. Virol J. 2017. PMID: 29100535 Free PMC article.
-
Determine the structure of phosphorylated modification of icariin and its antiviral activity against duck hepatitis virus A.BMC Vet Res. 2015 Aug 14;11:205. doi: 10.1186/s12917-015-0459-9. BMC Vet Res. 2015. PMID: 26272639 Free PMC article. Clinical Trial.
-
Widespread distribution and structural diversity of Type IV IRESs in members of Picornaviridae.Virology. 2015 Apr;478:61-74. doi: 10.1016/j.virol.2015.02.016. Epub 2015 Feb 27. Virology. 2015. PMID: 25726971 Free PMC article.
-
Cleavage of poly(A)-binding protein by duck hepatitis A virus 3C protease.Sci Rep. 2017 Nov 24;7(1):16261. doi: 10.1038/s41598-017-16484-1. Sci Rep. 2017. PMID: 29176600 Free PMC article.
References
-
- Belsham GJ. 2009. Divergent picornavirus IRES elements. Virus Res. 139:183–192 - PubMed
-
- Belsham GJ, Jackson RJ. 2000. Translation initiation on picornavirus RNA, p 869–900 In Sonenberg N, Hershey JWB, Mathews MB. (ed), Translational control of gene expression. Cold Spring Harbor monograph 39. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
-
- Bochkov YA, Palmenberg AC. 2006. Translational efficiency of EMCV IRES in bicistronic vectors is dependent upon IRES sequence and gene location. Biotechniques 41:283–284, 286, 288 passim - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

