Amino ester hydrolase from Xanthomonas campestris pv. campestris, ATCC 33913 for enzymatic synthesis of ampicillin
- PMID: 22087071
- PMCID: PMC3214638
- DOI: 10.1016/j.molcatb.2010.06.014
Amino ester hydrolase from Xanthomonas campestris pv. campestris, ATCC 33913 for enzymatic synthesis of ampicillin
Abstract
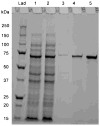
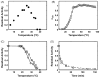
α-Amino ester hydrolases (AEH) are a small class of proteins, which are highly specific for hydrolysis or synthesis of α-amino containing amides and esters including β-lactam antibiotics such as ampicillin, amoxicillin, and cephalexin. A BLAST search revealed the sequence of a putative glutaryl 7-aminocephalosporanic acid (GL-7-ACA) acylase 93% identical to a known AEH from Xanthomonas citri. The gene, termed gaa, was cloned from the genomic DNA of Xanthomonas campestris pv. campestris sp. strain ATCC 33913 and the corresponding protein was expressed into Escherichia coli. The purified protein was able to perform both hydrolysis and synthesis of a variety of α-amino β-lactam antibiotics including (R)-ampicillin and cephalexin, with optimal ampicillin hydrolytic activity at 25 °C and pH 6.8, with kinetic parameters of k(cat) of 72.5 s(-1) and K(M) of 1.1 mM. The synthesis parameters α, β(o), and γ for ampicillin, determined here first for this class of proteins, are α = 0.25, β(o) = 42.8 M(-1), and γ = 0.23, and demonstrate the excellent synthetic potential of these enzymes. An extensive study of site-directed mutations around the binding pocket of X. campestris pv. campestris AEH strongly suggests that mutation of almost any first-shell amino acid residues around the active site leads to inactive enzyme, including Y82, Y175, D207, D208, W209, Y222, and E309, in addition to those residues forming the catalytic triad, S174, H340, and D307.
Figures

Similar articles
-
3D Structure Modeling of Alpha-Amino Acid Ester Hydrolase from Xanthomonas rubrilineans.Acta Naturae. 2013 Oct;5(4):62-70. Acta Naturae. 2013. PMID: 24455184 Free PMC article.
-
The sequence and crystal structure of the alpha-amino acid ester hydrolase from Xanthomonas citri define a new family of beta-lactam antibiotic acylases.J Biol Chem. 2003 Jun 20;278(25):23076-84. doi: 10.1074/jbc.M302246200. Epub 2003 Apr 8. J Biol Chem. 2003. PMID: 12684501
-
Cloning, sequence analysis, and expression in Escherichia coli of the gene encoding an alpha-amino acid ester hydrolase from Acetobacter turbidans.Appl Environ Microbiol. 2002 Jan;68(1):211-8. doi: 10.1128/AEM.68.1.211-218.2002. Appl Environ Microbiol. 2002. PMID: 11772629 Free PMC article.
-
[Purification and characterization of alpha-amino acid ester hydrolase from Xanthomonas rubrillineans].Wei Sheng Wu Xue Bao. 2012 May 4;52(5):620-8. Wei Sheng Wu Xue Bao. 2012. PMID: 22803348 Chinese.
-
Genetic organization of the lexA, recA and recX genes in Xanthomonas campestris.FEMS Microbiol Lett. 2002 Apr 9;209(2):149-54. doi: 10.1111/j.1574-6968.2002.tb11124.x. FEMS Microbiol Lett. 2002. PMID: 12007798
Cited by
-
Ampicillin Synthesis Using a Two-Enzyme Cascade with Both α-Amino Ester Hydrolase and Penicillin G Acylase.ChemCatChem. 2010 Aug 9;2(8):987-991. doi: 10.1002/cctc.201000135. ChemCatChem. 2010. PMID: 22039394 Free PMC article.
-
Improvement of α-amino Ester Hydrolase Stability via Computational Protein Design.Protein J. 2023 Dec;42(6):675-684. doi: 10.1007/s10930-023-10155-z. Epub 2023 Oct 11. Protein J. 2023. PMID: 37819423
-
Differentiation of Xylella fastidiosa strains via multilocus sequence analysis of environmentally mediated genes (MLSA-E).Appl Environ Microbiol. 2012 Mar;78(5):1385-96. doi: 10.1128/AEM.06679-11. Epub 2011 Dec 22. Appl Environ Microbiol. 2012. PMID: 22194287 Free PMC article.
-
Reactor Design and Optimization of α-Amino Ester Hydrolase- Catalyzed Synthesis of Cephalexin.Front Bioeng Biotechnol. 2022 Mar 2;10:826357. doi: 10.3389/fbioe.2022.826357. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 35309985 Free PMC article.
-
3D Structure Modeling of Alpha-Amino Acid Ester Hydrolase from Xanthomonas rubrilineans.Acta Naturae. 2013 Oct;5(4):62-70. Acta Naturae. 2013. PMID: 24455184 Free PMC article.
References
-
- Elander R. Appl Microbiol Biotechnol. 2003;61:385–392. - PubMed
-
- Deaguero AL, Blum JK, Bommarius AS. Wiley’s Encyclopedia of Industrial Biotechnology. In: Flickinger MC, editor. Biocatalytic Synthesis of Beta-Lactam Antibiotics. 2010. pp. 535–567.
-
- Alkema WBL, de Vries E, Floris R, Janssen DB. Eur J Biochem. 2003;270:3675–3683. - PubMed
-
- Arroyo M, de la Mata I, Acebal C, Castillon MP. Appl Microbiol Biotechnol. 2003;60:507–514. - PubMed
-
- Barends TRM, Polderman-Tijmes JJ, Jekel PA, Hensgens CMH, de Vries EJ, Janssen DB, Dijkstra BW. J Biol Chem. 2003;278:23076–23084. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
