Development of a 'clickable' non-natural nucleotide to visualize the replication of non-instructional DNA lesions
- PMID: 22086959
- PMCID: PMC3300027
- DOI: 10.1093/nar/gkr980
Development of a 'clickable' non-natural nucleotide to visualize the replication of non-instructional DNA lesions
Abstract
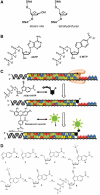
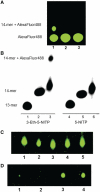
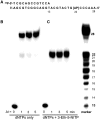
The misreplication of damaged DNA is an important biological process that produces numerous adverse effects on human health. This report describes the synthesis and characterization of a non-natural nucleotide, designated 3-ethynyl-5-nitroindolyl-2'-deoxyriboside triphosphate (3-Eth-5-NITP), as a novel chemical reagent that can probe and quantify the misreplication of damaged DNA. We demonstrate that this non-natural nucleotide is efficiently inserted opposite an abasic site, a commonly formed and potentially mutagenic non-instructional DNA lesion. The strategic placement of the ethynyl moiety allows the incorporated nucleoside triphosphate to be selectively tagged with an azide-containing fluorophore using 'click' chemistry. This reaction provides a facile way to quantify the extent of nucleotide incorporation opposite non-instructional DNA lesions. In addition, the incorporation of 3-Eth-5-NITP is highly selective for an abasic site, and occurs even in the presence of a 50-fold molar excess of natural nucleotides. The biological applications of using 3-Eth-5-NITP as a chemical probe to monitor and quantify the misreplication of non-instructional DNA lesions are discussed.
Figures

Similar articles
-
Artificial Nucleosides as Diagnostic Probes to Measure Translesion DNA Synthesis.Methods Mol Biol. 2019;1973:237-249. doi: 10.1007/978-1-4939-9216-4_15. Methods Mol Biol. 2019. PMID: 31016706
-
A non-natural nucleoside with combined therapeutic and diagnostic activities against leukemia.ACS Chem Biol. 2012 Jun 15;7(6):988-98. doi: 10.1021/cb300038f. Epub 2012 Mar 13. ACS Chem Biol. 2012. PMID: 22390204 Free PMC article.
-
Optimization of non-natural nucleotides for selective incorporation opposite damaged DNA.Org Biomol Chem. 2007 Nov 21;5(22):3623-30. doi: 10.1039/b712480e. Epub 2007 Oct 12. Org Biomol Chem. 2007. PMID: 17971991
-
Visualizing nucleic acid metabolism using non-natural nucleosides and nucleotide analogs.Biochim Biophys Acta. 2016 Jan;1864(1):165-76. doi: 10.1016/j.bbapap.2015.05.010. Epub 2015 May 22. Biochim Biophys Acta. 2016. PMID: 26004088 Review.
-
DNA Replication: From Radioisotopes to Click Chemistry.Molecules. 2018 Nov 17;23(11):3007. doi: 10.3390/molecules23113007. Molecules. 2018. PMID: 30453631 Free PMC article. Review.
Cited by
-
A Cascade of Thermophilic Enzymes As an Approach to the Synthesis of Modified Nucleotides.Acta Naturae. 2016 Oct-Dec;8(4):82-90. Acta Naturae. 2016. PMID: 28050269 Free PMC article.
-
Identification of DNA lesions using a third base pair for amplification and nanopore sequencing.Nat Commun. 2015 Nov 6;6:8807. doi: 10.1038/ncomms9807. Nat Commun. 2015. PMID: 26542210 Free PMC article.
-
Inhibiting translesion DNA synthesis as an approach to combat drug resistance to DNA damaging agents.Oncotarget. 2017 Jun 20;8(25):40804-40816. doi: 10.18632/oncotarget.17254. Oncotarget. 2017. PMID: 28489578 Free PMC article.
-
The use of modified and non-natural nucleotides provide unique insights into pro-mutagenic replication catalyzed by polymerase eta.Nucleic Acids Res. 2016 Feb 18;44(3):1022-35. doi: 10.1093/nar/gkv1509. Epub 2015 Dec 29. Nucleic Acids Res. 2016. PMID: 26717984 Free PMC article.
-
Inhibiting DNA Polymerases as a Therapeutic Intervention against Cancer.Front Mol Biosci. 2017 Nov 21;4:78. doi: 10.3389/fmolb.2017.00078. eCollection 2017. Front Mol Biosci. 2017. PMID: 29201867 Free PMC article. Review.
References
-
- Hanawalt PC. Paradigms for the three rs: DNA replication, recombination, and repair. Mol. Cell. 2007;28:702–707. - PubMed
-
- Pages V, Fuchs RP. How DNA lesions are turned into mutations within cells? Oncogene. 2002;21:8957–8966. - PubMed
-
- Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat. Res. 2005;577:55–76. - PubMed
-
- Nakamura J, Swenberg JA. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59:2522–2526. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

