Human tRNA genes function as chromatin insulators
- PMID: 22085927
- PMCID: PMC3261562
- DOI: 10.1038/emboj.2011.406
Human tRNA genes function as chromatin insulators
Abstract
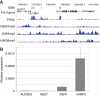
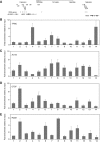
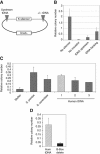
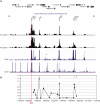
Insulators help separate active chromatin domains from silenced ones. In yeast, gene promoters act as insulators to block the spread of Sir and HP1 mediated silencing while in metazoans most insulators are multipartite autonomous entities. tDNAs are repetitive sequences dispersed throughout the human genome and we now show that some of these tDNAs can function as insulators in human cells. Using computational methods, we identified putative human tDNA insulators. Using silencer blocking, transgene protection and repressor blocking assays we show that some of these tDNA-containing fragments can function as barrier insulators in human cells. We find that these elements also have the ability to block enhancers from activating RNA pol II transcribed promoters. Characterization of a putative tDNA insulator in human cells reveals that the site possesses chromatin signatures similar to those observed at other better-characterized eukaryotic insulators. Enhanced 4C analysis demonstrates that the tDNA insulator makes long-range chromatin contacts with other tDNAs and ETC sites but not with intervening or flanking RNA pol II transcribed genes.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




Similar articles
-
tDNA insulators and the emerging role of TFIIIC in genome organization.Transcription. 2012 Nov-Dec;3(6):277-84. doi: 10.4161/trns.21579. Epub 2012 Aug 14. Transcription. 2012. PMID: 22889843 Free PMC article. Review.
-
TFIIIC-based chromatin insulators through eukaryotic evolution.Gene. 2022 Aug 15;835:146533. doi: 10.1016/j.gene.2022.146533. Epub 2022 May 24. Gene. 2022. PMID: 35623477
-
TFIIIC binding sites function as both heterochromatin barriers and chromatin insulators in Saccharomyces cerevisiae.Eukaryot Cell. 2008 Dec;7(12):2078-86. doi: 10.1128/EC.00128-08. Epub 2008 Oct 10. Eukaryot Cell. 2008. PMID: 18849469 Free PMC article.
-
TFIIIC bound DNA elements in nuclear organization and insulation.Biochim Biophys Acta. 2013 Mar-Apr;1829(3-4):418-24. doi: 10.1016/j.bbagrm.2012.09.006. Epub 2012 Sep 21. Biochim Biophys Acta. 2013. PMID: 23000638 Free PMC article. Review.
-
MIR retrotransposon sequences provide insulators to the human genome.Proc Natl Acad Sci U S A. 2015 Aug 11;112(32):E4428-37. doi: 10.1073/pnas.1507253112. Epub 2015 Jul 27. Proc Natl Acad Sci U S A. 2015. PMID: 26216945 Free PMC article.
Cited by
-
Silencing near tRNA genes is nucleosome-mediated and distinct from boundary element function.Gene. 2013 Aug 15;526(1):7-15. doi: 10.1016/j.gene.2013.05.016. Epub 2013 May 23. Gene. 2013. PMID: 23707796 Free PMC article.
-
tRNA processing defects induce replication stress and Chk2-dependent disruption of piRNA transcription.EMBO J. 2015 Dec 14;34(24):3009-27. doi: 10.15252/embj.201591006. Epub 2015 Oct 15. EMBO J. 2015. PMID: 26471728 Free PMC article.
-
Functional validation of a constitutive autonomous silencer element.PLoS One. 2015 Apr 24;10(4):e0124588. doi: 10.1371/journal.pone.0124588. eCollection 2015. PLoS One. 2015. PMID: 25910277 Free PMC article.
-
RNA polymerases reshape chromatin and coordinate transcription on individual fibers.bioRxiv [Preprint]. 2023 Dec 23:2023.12.22.573133. doi: 10.1101/2023.12.22.573133. bioRxiv. 2023. Update in: Mol Cell. 2024 Sep 5;84(17):3209-3222.e5. doi: 10.1016/j.molcel.2024.08.013. PMID: 38187631 Free PMC article. Updated. Preprint.
-
Poly(ADP-ribosyl)ation regulates insulator function and intrachromosomal interactions in Drosophila.Cell. 2013 Sep 26;155(1):148-59. doi: 10.1016/j.cell.2013.08.052. Epub 2013 Sep 19. Cell. 2013. PMID: 24055367 Free PMC article.
References
-
- Adams C, Haldar D, Kamakaka RT (2005) Construction and characterization of a series of vectors for Schizosaccharomyces pombe. Yeast 22: 1307–1314 - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
-
- Bartlett J, Blagojevic J, Carter D, Eskiw C, Fromaget M, Job C, Shamsher M, Trindade IF, Xu M, Cook PR (2006) Specialized transcription factories. Biochem Soc Symp 73: 67–75 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

