IGSF4 is a novel TCR ζ-chain-interacting protein that enhances TCR-mediated signaling
- PMID: 22084409
- PMCID: PMC3256964
- DOI: 10.1084/jem.20110853
IGSF4 is a novel TCR ζ-chain-interacting protein that enhances TCR-mediated signaling
Abstract
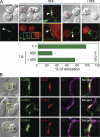
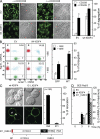
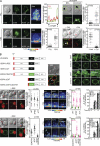
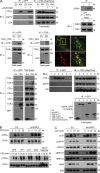
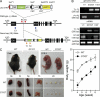
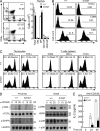
Immunoglobulin superfamily member 4 (IGSF4) is a known ligand of CRTAM, a receptor expressed in activated NKT and CD8(+) T cells, but its function in T cell immunity has not been elucidated. In this study, we show that IGSF4 directly interacts with the T cell receptor (TCR) ζ-chain and enhances TCR signaling by enhancing ζ-chain phosphorylation. Ectopic overexpression of IGSF4 enhances TCR-mediated T cell activation. In contrast, IGSF4 knockdown shows a dramatic decrease in markers associated with T cell activation compared with those in control small interfering RNA. The transmembrane domain is essential for TCR ζ-chain association and clustering to the immunological synapse, and the ectodomain is associated with T cell interaction with antigen-presenting cells (APCs). IGSF4-deficient mice have impaired TCR-mediated thymocyte selection and maturation. Furthermore, these mice reveal attenuated effector T cell functions accompanied by defective TCR signaling. Collectively, the results indicate that IGSF4 plays a central role in T cell functioning by dual independent mechanisms, control of TCR signaling and control of T cell-APC interaction.
Figures


Similar articles
-
Potentiating the Antitumor Activity of Cytotoxic T Cells via the Transmembrane Domain of IGSF4 That Increases TCR Avidity.Front Immunol. 2021 Feb 1;11:591054. doi: 10.3389/fimmu.2020.591054. eCollection 2020. Front Immunol. 2021. PMID: 33597944 Free PMC article.
-
Talin1 regulates TCR-mediated LFA-1 function.J Immunol. 2006 Dec 1;177(11):7707-14. doi: 10.4049/jimmunol.177.11.7707. J Immunol. 2006. PMID: 17114441
-
LFA-1-mediated T cell costimulation through increased localization of TCR/class II complexes to the central supramolecular activation cluster and exclusion of CD45 from the immunological synapse.J Immunol. 2007 Aug 1;179(3):1616-24. doi: 10.4049/jimmunol.179.3.1616. J Immunol. 2007. PMID: 17641028 Free PMC article.
-
Involvement of a cell adhesion molecule, TSLC1/IGSF4, in human oncogenesis.Cancer Sci. 2005 Sep;96(9):543-52. doi: 10.1111/j.1349-7006.2005.00089.x. Cancer Sci. 2005. PMID: 16128739 Free PMC article. Review.
-
Manipulating the TCR signaling network for cellular immunotherapy: Challenges & opportunities.Mol Immunol. 2020 Jul;123:64-73. doi: 10.1016/j.molimm.2020.04.007. Epub 2020 May 15. Mol Immunol. 2020. PMID: 32422416 Free PMC article. Review.
Cited by
-
IGSF1 deficiency syndrome: A newly uncovered endocrinopathy.Rare Dis. 2013 May 2;1:e24883. doi: 10.4161/rdis.24883. eCollection 2013. Rare Dis. 2013. PMID: 25002994 Free PMC article.
-
Nectin Family Ligands Trigger Immune Effector Functions in Health and Autoimmunity.Biology (Basel). 2023 Mar 15;12(3):452. doi: 10.3390/biology12030452. Biology (Basel). 2023. PMID: 36979144 Free PMC article. Review.
-
CRTAM determines the CD4+ cytotoxic T lymphocyte lineage.J Exp Med. 2016 Jan 11;213(1):123-38. doi: 10.1084/jem.20150519. Epub 2015 Dec 22. J Exp Med. 2016. PMID: 26694968 Free PMC article.
-
CADM1 is essential for KSHV-encoded vGPCR-and vFLIP-mediated chronic NF-κB activation.PLoS Pathog. 2018 Apr 26;14(4):e1006968. doi: 10.1371/journal.ppat.1006968. eCollection 2018 Apr. PLoS Pathog. 2018. PMID: 29698475 Free PMC article.
-
Human T-cell leukemia virus type 1 (HTLV-1) tax requires CADM1/TSLC1 for inactivation of the NF-κB inhibitor A20 and constitutive NF-κB signaling.PLoS Pathog. 2015 Mar 16;11(3):e1004721. doi: 10.1371/journal.ppat.1004721. eCollection 2015 Mar. PLoS Pathog. 2015. PMID: 25774694 Free PMC article.
References
-
- Abbas A.R., Baldwin D., Ma Y., Ouyang W., Gurney A., Martin F., Fong S., van Lookeren Campagne M., Godowski P., Williams P.M., et al. 2005. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 6:319–331 10.1038/sj.gene.6364173 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

