Reduced DNA polytenization of a minichromosome region undergoing position-effect variegation in Drosophila
- PMID: 2208283
- PMCID: PMC3229194
- DOI: 10.1016/0092-8674(90)90291-l
Reduced DNA polytenization of a minichromosome region undergoing position-effect variegation in Drosophila
Abstract
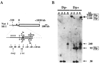
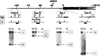
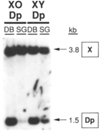
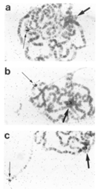
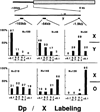
Molecular analysis of a Drosophila minichromosome, Dp(1;f)1187, revealed a relationship between position-effect variegation and the copy number reductions of heterochromatic sequences that occur in polytene cells. Heterochromatin adjacent to a defined junction with euchromatin underpolytenized at least 60-fold. Lesser reductions were observed in euchromatic sequences up to 103 kb from the breakpoint. The copy number changes behaved in all respects like the expression of yellow, a gene located within the affected region. Both copy number and yellow expression displayed a cell-by-cell mosaic pattern of reduction, and adding a Y chromosome, a known suppressor of variegation, increased both substantially. We discuss the possibility that changes in replication alter copy number locally and also propose an alternative model of position-effect variegation based on the somatic elimination of heterochromatic sequences.
Figures

Similar articles
-
Replication forks are not found in a Drosophila minichromosome demonstrating a gradient of polytenization.Chromosoma. 1992 Dec;102(1):15-9. doi: 10.1007/BF00352285. Chromosoma. 1992. PMID: 1291225
-
Replication of heterochromatin and structure of polytene chromosomes.Mol Cell Biol. 2000 Sep;20(17):6308-16. doi: 10.1128/MCB.20.17.6308-6316.2000. Mol Cell Biol. 2000. PMID: 10938107 Free PMC article.
-
Unusual properties of genomic DNA molecules spanning the euchromatic-heterochromatic junction of a Drosophila minichromosome.Nucleic Acids Res. 1994 Nov 25;22(23):5068-75. doi: 10.1093/nar/22.23.5068. Nucleic Acids Res. 1994. PMID: 7800501 Free PMC article.
-
Chromatin structure and the regulation of gene expression: the lessons of PEV in Drosophila.Adv Genet. 2008;61:1-43. doi: 10.1016/S0065-2660(07)00001-6. Adv Genet. 2008. PMID: 18282501 Review.
-
[Distorted heterochromatin replication in Drosophila melanogaster polytene chromosomes as a result of euchromatin-heterochromatin rearrangements].Genetika. 2007 Jan;43(1):18-26. Genetika. 2007. PMID: 17333934 Review. Russian.
Cited by
-
Distinct families of site-specific retrotransposons occupy identical positions in the rRNA genes of Anopheles gambiae.Mol Cell Biol. 1992 Nov;12(11):5102-10. doi: 10.1128/mcb.12.11.5102-5110.1992. Mol Cell Biol. 1992. PMID: 1328871 Free PMC article.
-
A sex-influenced modifier in Drosophila that affects a broad spectrum of target loci including the histone repeats.Genetics. 1997 Jul;146(3):903-17. doi: 10.1093/genetics/146.3.903. Genetics. 1997. PMID: 9215896 Free PMC article.
-
Cis-effects of heterochromatin on heterochromatic and euchromatic gene activity in Drosophila melanogaster.Genetics. 1995 Jul;140(3):1033-45. doi: 10.1093/genetics/140.3.1033. Genetics. 1995. PMID: 7672575 Free PMC article.
-
Molecular structure of a functional Drosophila centromere.Cell. 1997 Dec 26;91(7):1007-19. doi: 10.1016/s0092-8674(00)80491-2. Cell. 1997. PMID: 9428523 Free PMC article.
-
SU(VAR)3-7, a Drosophila heterochromatin-associated protein and companion of HP1 in the genomic silencing of position-effect variegation.EMBO J. 1997 Sep 1;16(17):5280-8. doi: 10.1093/emboj/16.17.5280. EMBO J. 1997. PMID: 9311988 Free PMC article.
References
-
- Ananiev EV, Gvozdev VA. Changed pattern of transcription and replication in polytene chromosomes of Drosophila resulting from eu-heterochromatin rearrangement. Chromosoma. 1974;45:173–191. - PubMed
-
- Ashburner M. Drosophila: A Laboratory Handbook. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1990. p. 1331.
-
- Beckingham K, Rubacha A. Different chromatin states of the intron− and type 1 intron+ rRNA genes of Calliphora erythrocephela. Chromosoma. 1984;90:311–316. - PubMed
-
- Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. - PubMed
-
- Bryan GJ, Jacobson JW, Hartl DL. Heritable somatic excision of a Drosophila transposon. Science. 1987;235:1636–1638. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

