Tim50's presequence receptor domain is essential for signal driven transport across the TIM23 complex
- PMID: 22065641
- PMCID: PMC3257539
- DOI: 10.1083/jcb.201105098
Tim50's presequence receptor domain is essential for signal driven transport across the TIM23 complex
Abstract
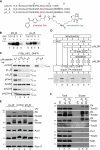
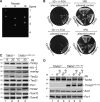
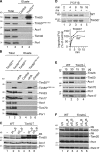
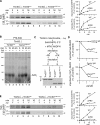
N-terminal targeting signals (presequences) direct proteins across the TOM complex in the outer mitochondrial membrane and the TIM23 complex in the inner mitochondrial membrane. Presequences provide directionality to the transport process and regulate the transport machineries during translocation. However, surprisingly little is known about how presequence receptors interact with the signals and what role these interactions play during preprotein transport. Here, we identify signal-binding sites of presequence receptors through photo-affinity labeling. Using engineered presequence probes, photo cross-linking sites on mitochondrial proteins were mapped mass spectrometrically, thereby defining a presequence-binding domain of Tim50, a core subunit of the TIM23 complex that is essential for mitochondrial protein import. Our results establish Tim50 as the primary presequence receptor at the inner membrane and show that targeting signals and Tim50 regulate the Tim23 channel in an antagonistic manner.
© 2011 Schulz et al.
Figures


Similar articles
-
Transmembrane Coordination of Preprotein Recognition and Motor Coupling by the Mitochondrial Presequence Receptor Tim50.Cell Rep. 2020 Mar 3;30(9):3092-3104.e4. doi: 10.1016/j.celrep.2020.02.031. Cell Rep. 2020. PMID: 32130909
-
Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes.Cell. 2002 Nov 15;111(4):519-28. doi: 10.1016/s0092-8674(02)01053-x. Cell. 2002. PMID: 12437925
-
Mgr2 promotes coupling of the mitochondrial presequence translocase to partner complexes.J Cell Biol. 2012 May 28;197(5):595-604. doi: 10.1083/jcb.201110047. Epub 2012 May 21. J Cell Biol. 2012. PMID: 22613836 Free PMC article.
-
On the mechanism of preprotein import by the mitochondrial presequence translocase.Biochim Biophys Acta. 2010 Jun;1803(6):732-9. doi: 10.1016/j.bbamcr.2010.01.013. Epub 2010 Jan 25. Biochim Biophys Acta. 2010. PMID: 20100523 Review.
-
From TOM to the TIM23 complex - handing over of a precursor.Biol Chem. 2020 May 26;401(6-7):709-721. doi: 10.1515/hsz-2020-0101. Biol Chem. 2020. PMID: 32074073 Review.
Cited by
-
Protein import motor complex reacts to mitochondrial misfolding by reducing protein import and activating mitophagy.Nat Commun. 2022 Sep 2;13(1):5164. doi: 10.1038/s41467-022-32564-x. Nat Commun. 2022. PMID: 36056001 Free PMC article.
-
Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution.Nat Struct Mol Biol. 2019 Dec;26(12):1158-1166. doi: 10.1038/s41594-019-0339-2. Epub 2019 Nov 18. Nat Struct Mol Biol. 2019. PMID: 31740857 Free PMC article.
-
Long Non-coding RNAs With In Vitro and In Vivo Efficacy in Preclinical Models of Esophageal Squamous Cell Carcinoma Which Act by a Non-microRNA Sponging Mechanism.Cancer Genomics Proteomics. 2022 Jul-Aug;19(4):372-389. doi: 10.21873/cgp.20327. Cancer Genomics Proteomics. 2022. PMID: 35732324 Free PMC article. Review.
-
Cation selectivity of the presequence translocase channel Tim23 is crucial for efficient protein import.Elife. 2017 Aug 31;6:e28324. doi: 10.7554/eLife.28324. Elife. 2017. PMID: 28857742 Free PMC article.
-
ROMO1 is a constituent of the human presequence translocase required for YME1L protease import.J Cell Biol. 2019 Feb 4;218(2):598-614. doi: 10.1083/jcb.201806093. Epub 2018 Dec 31. J Cell Biol. 2019. PMID: 30598479 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

