An integrated transcriptomic and meta-analysis of hepatoma cells reveals factors that influence susceptibility to HCV infection
- PMID: 22046242
- PMCID: PMC3201949
- DOI: 10.1371/journal.pone.0025584
An integrated transcriptomic and meta-analysis of hepatoma cells reveals factors that influence susceptibility to HCV infection
Abstract
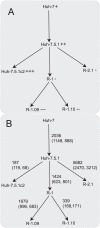
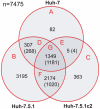
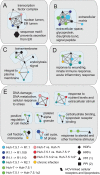
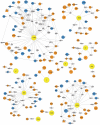
Hepatitis C virus (HCV) is a global problem. To better understand HCV infection researchers employ in vitro HCV cell-culture (HCVcc) systems that use Huh-7 derived hepatoma cells that are particularly permissive to HCV infection. A variety of hyper-permissive cells have been subcloned for this purpose. In addition, subclones of Huh-7 which have evolved resistance to HCV are available. However, the mechanisms of susceptibility or resistance to infection among these cells have not been fully determined. In order to elucidate mechanisms by which hepatoma cells are susceptible or resistant to HCV infection we performed genome-wide expression analyses of six Huh-7 derived cell cultures that have different levels of permissiveness to infection. A great number of genes, representing a wide spectrum of functions are differentially expressed between cells. To focus our investigation, we identify host proteins from HCV replicase complexes, perform gene expression analysis of three HCV infected cells and conduct a detailed analysis of differentially expressed host factors by integrating a variety of data sources. Our results demonstrate that changes relating to susceptibility to HCV infection in hepatoma cells are linked to the innate immune response, secreted signal peptides and host factors that have a role in virus entry and replication. This work identifies both known and novel host factors that may influence HCV infection. Our findings build upon current knowledge of the complex interplay between HCV and the host cell, which could aid development of new antiviral strategies.
Conflict of interest statement
Figures



Similar articles
-
HepG2 cells mount an effective antiviral interferon-lambda based innate immune response to hepatitis C virus infection.Hepatology. 2014 Oct;60(4):1170-9. doi: 10.1002/hep.27227. Epub 2014 Aug 21. Hepatology. 2014. PMID: 24833036 Free PMC article.
-
Permissiveness of human hepatocellular carcinoma cell lines for hepatitis C virus entry and replication.Virus Res. 2017 Aug 15;240:35-46. doi: 10.1016/j.virusres.2017.07.018. Epub 2017 Jul 24. Virus Res. 2017. PMID: 28751105
-
Hepatitis C virus triggers apoptosis of a newly developed hepatoma cell line through antiviral defense system.Gastroenterology. 2007 Nov;133(5):1649-59. doi: 10.1053/j.gastro.2007.09.017. Epub 2007 Sep 16. Gastroenterology. 2007. PMID: 17983809
-
Interferon Response in Hepatitis C Virus (HCV) Infection: Lessons from Cell Culture Systems of HCV Infection.Int J Mol Sci. 2015 Oct 7;16(10):23683-94. doi: 10.3390/ijms161023683. Int J Mol Sci. 2015. PMID: 26457705 Free PMC article. Review.
-
Immunogenetic studies of the hepatitis C virus infection in an era of pan-genotype antiviral therapies - Effective treatment is coming.Infect Genet Evol. 2018 Dec;66:376-391. doi: 10.1016/j.meegid.2017.08.011. Epub 2017 Aug 12. Infect Genet Evol. 2018. PMID: 28811194 Review.
Cited by
-
Dual Effects of Let-7b in the Early Stage of Hepatitis C Virus Infection.J Virol. 2021 Jan 28;95(4):e01800-20. doi: 10.1128/JVI.01800-20. Print 2021 Jan 28. J Virol. 2021. PMID: 33208444 Free PMC article.
-
Retracted Article: The synthesis and biological activity of marine alkaloid derivatives and analogues.RSC Adv. 2020 Aug 28;10(53):31909-31935. doi: 10.1039/d0ra05856d. eCollection 2020 Aug 26. RSC Adv. 2020. Retraction in: RSC Adv. 2021 Feb 22;11(14):7896. doi: 10.1039/d1ra90083h PMID: 35518151 Free PMC article. Retracted.
-
The lipid kinase phosphatidylinositol-4 kinase III alpha regulates the phosphorylation status of hepatitis C virus NS5A.PLoS Pathog. 2013 May;9(5):e1003359. doi: 10.1371/journal.ppat.1003359. Epub 2013 May 9. PLoS Pathog. 2013. PMID: 23675303 Free PMC article.
-
RNA structural elements of hepatitis C virus controlling viral RNA translation and the implications for viral pathogenesis.Viruses. 2012 Oct 19;4(10):2233-50. doi: 10.3390/v4102233. Viruses. 2012. PMID: 23202462 Free PMC article. Review.
-
Identification of HNRNPK as regulator of hepatitis C virus particle production.PLoS Pathog. 2015 Jan 8;11(1):e1004573. doi: 10.1371/journal.ppat.1004573. eCollection 2015 Jan. PLoS Pathog. 2015. PMID: 25569684 Free PMC article.
References
-
- Frank C, Mohamed M, Strickland G, Lavanchy D, Arthur R, et al. The role of parenteral antischistosomal therapy in the spread of hepatitis C virus in Egypt. The Lancet. 2000;355:887–891. - PubMed
-
- Pawlotsky J. Therapy of hepatitis C: from empiricism to eradication. Hepatology. 2006;43:S207–S220. - PubMed
-
- Lemon S, McKeating J, Pietschmann T, Frick D, Glenn J, et al. Development of novel therapies for hepatitis C. Antiviral Research. 2010;86:79–92. - PubMed
-
- Coelmont L, Kaptein S, Paeshuyse J, Vliegen I, Dumont J, et al. Debio 025, a cyclophilin binding molecule, is highly efficient in clearing hepatitis C virus (HCV) replicon-containing cells when used alone or in combination with specifically targeted antiviral therapy for HCV (STAT-C) inhibitors. Antimicrobial agents and chemotherapy. 2009;53:967. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

