In vitro repair of DNA hairpins containing various numbers of CAG/CTG trinucleotide repeats
- PMID: 22041023
- PMCID: PMC3356785
- DOI: 10.1016/j.dnarep.2011.10.020
In vitro repair of DNA hairpins containing various numbers of CAG/CTG trinucleotide repeats
Abstract
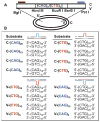
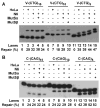
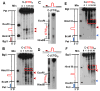
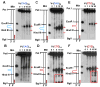
Expansion of CAG/CTG trinucleotide repeats (TNRs) in humans is associated with a number of neurological and neurodegenerative disorders including Huntington's disease. Increasing evidence suggests that formation of a stable DNA hairpin within CAG/CTG repeats during DNA metabolism leads to TNR instability. However, the molecular mechanism by which cells recognize and repair CAG/CTG hairpins is largely unknown. Recent studies have identified a novel DNA repair pathway specifically removing (CAG)(n)/(CTG)(n) hairpins, which is considered a major mechanism responsible for TNR instability. The hairpin repair (HPR) system targets the repeat tracts for incisions in the nicked strand in an error-free manner. To determine the substrate spectrum of the HPR system and its ability to process smaller hairpins, which may be the intermediates for CAG/CTG expansions, we constructed a series of CAG/CTG hairpin heteroduplexes containing different numbers of repeats (from 5 to 25) and examined their repair in human nuclear extracts. We show here that although repair efficiencies differ slightly among these substrates, removal of the individual hairpin structures all involve endonucleolytic incisions within the repeat tracts in the nicked DNA strand. Analysis of the repair intermediates defined specific incision sites for each substrate, which were all located within the repeat regions. Mismatch repair proteins are not required for, nor do they inhibit, the processing of smaller hairpin structures. These results suggest that the HPR system ensures CAG/CTG stability primarily by removing various sizes of (CAG)(n)/(CTG)(n) hairpin structures during DNA metabolism.
Copyright © 2011 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



Similar articles
-
Incision-dependent and error-free repair of (CAG)(n)/(CTG)(n) hairpins in human cell extracts.Nat Struct Mol Biol. 2009 Aug;16(8):869-75. doi: 10.1038/nsmb.1638. Epub 2009 Jul 13. Nat Struct Mol Biol. 2009. PMID: 19597480 Free PMC article.
-
The Werner syndrome protein promotes CAG/CTG repeat stability by resolving large (CAG)(n)/(CTG)(n) hairpins.J Biol Chem. 2012 Aug 31;287(36):30151-6. doi: 10.1074/jbc.M112.389791. Epub 2012 Jul 11. J Biol Chem. 2012. PMID: 22787159 Free PMC article.
-
The Role of XPG in Processing (CAG)n/(CTG)n DNA Hairpins.Cell Biosci. 2011 Mar 9;1(1):11. doi: 10.1186/2045-3701-1-11. Cell Biosci. 2011. PMID: 21711735 Free PMC article.
-
Structural and Dynamical Properties of Nucleic Acid Hairpins Implicated in Trinucleotide Repeat Expansion Diseases.Biomolecules. 2024 Oct 10;14(10):1278. doi: 10.3390/biom14101278. Biomolecules. 2024. PMID: 39456210 Free PMC article. Review.
-
Disease-associated repeat instability and mismatch repair.DNA Repair (Amst). 2016 Feb;38:117-126. doi: 10.1016/j.dnarep.2015.11.008. Epub 2015 Dec 12. DNA Repair (Amst). 2016. PMID: 26774442 Review.
Cited by
-
Coordinated processing of 3' slipped (CAG)n/(CTG)n hairpins by DNA polymerases β and δ preferentially induces repeat expansions.J Biol Chem. 2013 May 24;288(21):15015-22. doi: 10.1074/jbc.M113.464370. Epub 2013 Apr 12. J Biol Chem. 2013. PMID: 23585564 Free PMC article.
-
Modifiers of CAG/CTG Repeat Instability: Insights from Mammalian Models.J Huntingtons Dis. 2021;10(1):123-148. doi: 10.3233/JHD-200426. J Huntingtons Dis. 2021. PMID: 33579861 Free PMC article. Review.
-
The Repeat Expansion Diseases: The dark side of DNA repair.DNA Repair (Amst). 2015 Aug;32:96-105. doi: 10.1016/j.dnarep.2015.04.019. Epub 2015 Apr 30. DNA Repair (Amst). 2015. PMID: 26002199 Free PMC article. Review.
-
The dual nature of mismatch repair as antimutator and mutator: for better or for worse.Front Genet. 2014 Aug 21;5:287. doi: 10.3389/fgene.2014.00287. eCollection 2014. Front Genet. 2014. PMID: 25191341 Free PMC article. Review.
-
HDAC3 deacetylates the DNA mismatch repair factor MutSβ to stimulate triplet repeat expansions.Proc Natl Acad Sci U S A. 2020 Sep 22;117(38):23597-23605. doi: 10.1073/pnas.2013223117. Epub 2020 Sep 8. Proc Natl Acad Sci U S A. 2020. PMID: 32900932 Free PMC article.
References
-
- Lopez Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol. 2010;11:165–170. - PubMed
-
- Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. - PubMed
-
- Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. - PubMed
-
- Gordenin DA, Kunkel TA, Resnick MA. Repeat expansion—all in a flap? Nat Genet. 1997;16:116–118. - PubMed
-
- Kang S, Jaworski A, Ohshima K, Wells RD. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat Genet. 1995;10:213–218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

