Automatic rebuilding and optimization of crystallographic structures in the Protein Data Bank
- PMID: 22034521
- PMCID: PMC3232375
- DOI: 10.1093/bioinformatics/btr590
Automatic rebuilding and optimization of crystallographic structures in the Protein Data Bank
Abstract
Motivation: Macromolecular crystal structures in the Protein Data Bank (PDB) are a key source of structural insight into biological processes. These structures, some >30 years old, were constructed with methods of their era. With PDB_REDO, we aim to automatically optimize these structures to better fit their corresponding experimental data, passing the benefits of new methods in crystallography on to a wide base of non-crystallographer structure users.
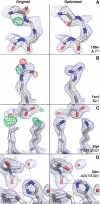
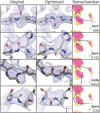
Results: We developed new algorithms to allow automatic rebuilding and remodeling of main chain peptide bonds and side chains in crystallographic electron density maps, and incorporated these and further enhancements in the PDB_REDO procedure. Applying the updated PDB_REDO to the oldest, but also to some of the newest models in the PDB, corrects existing modeling errors and brings these models to a higher quality, as judged by standard validation methods.
Availability and implementation: The PDB_REDO database and links to all software are available at http://www.cmbi.ru.nl/pdb_redo.
Contact: r.joosten@nki.nl; a.perrakis@nki.nl
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures

Similar articles
-
A series of PDB-related databanks for everyday needs.Nucleic Acids Res. 2015 Jan;43(Database issue):D364-8. doi: 10.1093/nar/gku1028. Epub 2014 Oct 28. Nucleic Acids Res. 2015. PMID: 25352545 Free PMC article.
-
PDB_REDO: constructive validation, more than just looking for errors.Acta Crystallogr D Biol Crystallogr. 2012 Apr;68(Pt 4):484-96. doi: 10.1107/S0907444911054515. Epub 2012 Mar 16. Acta Crystallogr D Biol Crystallogr. 2012. PMID: 22505269 Free PMC article.
-
Homology-based hydrogen bond information improves crystallographic structures in the PDB.Protein Sci. 2018 Mar;27(3):798-808. doi: 10.1002/pro.3353. Epub 2017 Dec 8. Protein Sci. 2018. PMID: 29168245 Free PMC article.
-
New Biological Insights from Better Structure Models.J Mol Biol. 2016 Mar 27;428(6):1375-1393. doi: 10.1016/j.jmb.2016.02.002. Epub 2016 Feb 8. J Mol Biol. 2016. PMID: 26869101 Review.
-
Integrative/Hybrid Methods Structural Biology: Role of Macromolecular Crystallography.Adv Exp Med Biol. 2018;1105:11-18. doi: 10.1007/978-981-13-2200-6_2. Adv Exp Med Biol. 2018. PMID: 30617820 Review.
Cited by
-
Structure of human factor VIIa-soluble tissue factor with calcium, magnesium and rubidium.Acta Crystallogr D Struct Biol. 2021 Jun 1;77(Pt 6):809-819. doi: 10.1107/S2059798321003922. Epub 2021 May 14. Acta Crystallogr D Struct Biol. 2021. PMID: 34076594 Free PMC article.
-
Protein Structure Idealization: How accurately is it possible to model protein structures with dihedral angles?Algorithms Mol Biol. 2013 Feb 25;8(1):5. doi: 10.1186/1748-7188-8-5. Algorithms Mol Biol. 2013. PMID: 23442792 Free PMC article.
-
Anomalies in the refinement of isoleucine.Acta Crystallogr D Biol Crystallogr. 2014 Apr;70(Pt 4):1037-49. doi: 10.1107/S139900471400087X. Epub 2014 Mar 19. Acta Crystallogr D Biol Crystallogr. 2014. PMID: 24699648 Free PMC article.
-
A series of PDB-related databanks for everyday needs.Nucleic Acids Res. 2015 Jan;43(Database issue):D364-8. doi: 10.1093/nar/gku1028. Epub 2014 Oct 28. Nucleic Acids Res. 2015. PMID: 25352545 Free PMC article.
-
Databases, Repositories, and Other Data Resources in Structural Biology.Methods Mol Biol. 2017;1607:643-665. doi: 10.1007/978-1-4939-7000-1_27. Methods Mol Biol. 2017. PMID: 28573593 Free PMC article. Review.
References
-
- Bernstein F.C., et al. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. - PubMed
-
- Cohen S.X., et al. Towards complete validated models in the next generation of ARP/wARP. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2222–2229. - PubMed
-
- Cowtan K. The Clipper C++ libraries for x-ray crystallography. IUCr Comput. Comission Newsl. 2003;2:4–9.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

