The metabolic regulator ERRα, a downstream target of HER2/IGF-1R, as a therapeutic target in breast cancer
- PMID: 22014575
- PMCID: PMC3199323
- DOI: 10.1016/j.ccr.2011.08.023
The metabolic regulator ERRα, a downstream target of HER2/IGF-1R, as a therapeutic target in breast cancer
Abstract
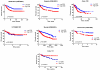
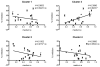
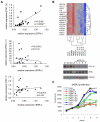
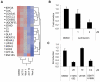
A genomic signature designed to assess the activity of the estrogen-related receptor alpha (ERRα) was used to profile more than 800 breast tumors, revealing a shorter disease-free survival in patients with tumors exhibiting elevated receptor activity. Importantly, this signature also predicted the ability of an ERRα antagonist, XCT790, to inhibit proliferation in cellular models of breast cancer. Using a chemical genomic approach, it was determined that activation of the Her2/IGF-1R signaling pathways and subsequent C-MYC stabilization upregulate the expression of peroxisome proliferator-activated receptor gamma coactivator-1 beta (PGC-1β), an obligate cofactor for ERRα activity. PGC-1β knockdown in breast cancer cells impaired ERRα signaling and reduced cell proliferation, implicating a functional role for PGC-1β/ERRα in the pathogenesis of breast cancers.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
PGC-1β regulates HER2-overexpressing breast cancer cells proliferation by metabolic and redox pathways.Tumour Biol. 2016 May;37(5):6035-44. doi: 10.1007/s13277-015-4449-0. Epub 2015 Nov 24. Tumour Biol. 2016. PMID: 26602383
-
Regulation of PPARgamma coactivator 1alpha (PGC-1alpha) signaling by an estrogen-related receptor alpha (ERRalpha) ligand.Proc Natl Acad Sci U S A. 2004 Jun 15;101(24):8912-7. doi: 10.1073/pnas.0401420101. Epub 2004 Jun 7. Proc Natl Acad Sci U S A. 2004. PMID: 15184675 Free PMC article.
-
[XCT790 inhibits rat vascular smooth muscle cells proliferation through down-regulating the expression of estrogen-related receptor alpha].Yao Xue Xue Bao. 2014 Feb;49(2):190-7. Yao Xue Xue Bao. 2014. PMID: 24761608 Chinese.
-
Molecular pathways: the metabolic regulator estrogen-related receptor α as a therapeutic target in cancer.Clin Cancer Res. 2012 Nov 15;18(22):6089-95. doi: 10.1158/1078-0432.CCR-11-3221. Epub 2012 Sep 27. Clin Cancer Res. 2012. PMID: 23019305 Free PMC article.
-
Estrogen-related receptor α regulates skeletal myocyte differentiation via modulation of the ERK MAP kinase pathway.Am J Physiol Cell Physiol. 2011 Sep;301(3):C630-45. doi: 10.1152/ajpcell.00033.2011. Epub 2011 May 11. Am J Physiol Cell Physiol. 2011. PMID: 21562305 Free PMC article.
Cited by
-
Inhibition of DNMT1 and ERRα crosstalk suppresses breast cancer via derepression of IRF4.Oncogene. 2020 Oct;39(41):6406-6420. doi: 10.1038/s41388-020-01438-1. Epub 2020 Aug 27. Oncogene. 2020. PMID: 32855526 Free PMC article.
-
Cholesterol and Mevalonate: Two Metabolites Involved in Breast Cancer Progression and Drug Resistance through the ERRα Pathway.Cells. 2020 Jul 31;9(8):1819. doi: 10.3390/cells9081819. Cells. 2020. PMID: 32751976 Free PMC article.
-
Estrogen-Related Receptor Alpha (ERRα) Promotes Cancer Stem Cell-Like Characteristics in Breast Cancer.Stem Cell Rev Rep. 2023 Nov;19(8):2807-2819. doi: 10.1007/s12015-023-10605-2. Epub 2023 Aug 16. Stem Cell Rev Rep. 2023. PMID: 37584854
-
Estrogen-related receptor α decreases RHOA stability to induce orientated cell migration.Proc Natl Acad Sci U S A. 2014 Oct 21;111(42):15108-13. doi: 10.1073/pnas.1402094111. Epub 2014 Oct 6. Proc Natl Acad Sci U S A. 2014. PMID: 25288732 Free PMC article.
-
Perturbations of cancer cell metabolism by the antidiabetic drug canagliflozin.Neoplasia. 2021 Apr;23(4):391-399. doi: 10.1016/j.neo.2021.02.003. Epub 2021 Mar 27. Neoplasia. 2021. PMID: 33784591 Free PMC article.
References
-
- Arany Z, Foo SY, Ma Y, Ruas JL, Bommi-Reddy A, Girnun G, Cooper M, Laznik D, Chinsomboon J, Rangwala SM, et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature. 2008;451:1008–1012. - PubMed
-
- Ariazi EA, Clark GM, Mertz JE. Estrogen-related receptor alpha and estrogen-related receptor gamma associate with unfavorable and favorable biomarkers, respectively, in human breast cancer. Cancer Res. 2002;62:6510–6518. - PubMed
-
- Ariazi EA, Kraus RJ, Farrell ML, Jordan VC, Mertz JE. Estrogen-related receptor alpha1 transcriptional activities are regulated in part via the ErbB2/HER2 signaling pathway. Mol Cancer Res. 2007;5:71–85. - PubMed
-
- Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

