Hypomethylation of the hsa-miR-191 locus causes high expression of hsa-mir-191 and promotes the epithelial-to-mesenchymal transition in hepatocellular carcinoma
- PMID: 21969817
- PMCID: PMC3182276
- DOI: 10.1593/neo.11698
Hypomethylation of the hsa-miR-191 locus causes high expression of hsa-mir-191 and promotes the epithelial-to-mesenchymal transition in hepatocellular carcinoma
Abstract
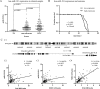
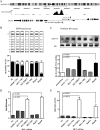
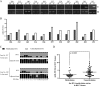
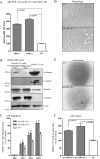
hsa-miR-191 is highly expressed in hepatocellular carcinoma (HCC), but the factors regulating this elevated expression are unknown. This study aimed to investigate the epigenetic mechanisms of increased hsa-miR-191 expression by analyzing the relationship between the DNA methylation status of hsa-miR-191 and miR-191 expression. Methylation-specific polymerase chain reaction (PCR), bisulfite sequencing PCR, Northern blot, and quantitative real-time PCR were performed to examine hsa-miR-191 methylation and expression levels. Western blot, transwell, and scratch assays were performed to examine the function and molecular mechanisms of hsa-miR-191. Approximately 58.9% of hsa-miR-191 expression was higher in HCC tissues than in adjacent noncancerous tissues; this high expression was associated with poor prognosis. The hypomethylation observed in some HCC cell lines and HCC tissues was correlated with the hsa-miR-191 expression level. This correlation was validated by treatment with the 5-aza-DAC demethylation agent. The level of hypomethylation was 63.0% in 73 clinical HCC tissue samples and was associated with increased (2.1-fold) hsa-miR-191 expression. The elevated expression of hsa-miR-191 in the SMMC-771 HCC cell line induced the cells to transition into mesenchymal-like cells; they exhibited characteristics such as loss of adhesion, down-regulation of epithelial cell markers, up-regulation of mesenchymal cell markers, and increased cell migration and invasion. Inhibiting hsa-miR-191 expression in the SMMC-7721 cell line reversed this process (as assessed by cell morphology and cell markers). Furthermore, hsa-miR-191 probably exerted its function by directly targeting TIMP metallopeptidase inhibitor 3 and inhibiting TIMP3 protein expression. Our results suggest that hsa-miR-191 locus hypomethylation causes an increase in hsa-miR-191 expression in HCC clinical tissues and that this expression induces HCC cells to transition into mesenchymal-like cells.
Figures



Similar articles
-
MicroRNA-370 Regulates Cellepithelial-Mesenchymal Transition, Migration, Invasion, and Prognosis of Hepatocellular Carcinoma by Targeting GUCD1.Yonsei Med J. 2019 Mar;60(3):267-276. doi: 10.3349/ymj.2019.60.3.267. Yonsei Med J. 2019. PMID: 30799589 Free PMC article.
-
[The expression and clinopathological significance of miR-130b in human hepatocellular carcinoma].Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2016 Mar;32(3):387-92. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2016. PMID: 26927562 Chinese.
-
miR-300 regulates the epithelial-mesenchymal transition and invasion of hepatocellular carcinoma by targeting the FAK/PI3K/AKT signaling pathway.Biomed Pharmacother. 2018 Jul;103:1632-1642. doi: 10.1016/j.biopha.2018.03.005. Epub 2018 May 7. Biomed Pharmacother. 2018. PMID: 29864952
-
Antitumor function of microRNA-122 against hepatocellular carcinoma.J Gastroenterol. 2014 Apr;49(4):589-93. doi: 10.1007/s00535-014-0932-4. Epub 2014 Feb 17. J Gastroenterol. 2014. PMID: 24531873 Review.
-
Specific molecular signatures of non-tumor liver tissue may predict a risk of hepatocarcinogenesis.Cancer Sci. 2014 Jul;105(7):749-54. doi: 10.1111/cas.12431. Epub 2014 Jun 18. Cancer Sci. 2014. PMID: 24766251 Free PMC article. Review.
Cited by
-
Serum microRNAs as biomarkers for recurrence in melanoma.J Transl Med. 2012 Aug 2;10:155. doi: 10.1186/1479-5876-10-155. J Transl Med. 2012. PMID: 22857597 Free PMC article.
-
miR-23b as a potential tumor suppressor and its regulation by DNA methylation in cervical cancer.Infect Agent Cancer. 2015 Nov 30;10:42. doi: 10.1186/s13027-015-0037-6. eCollection 2015. Infect Agent Cancer. 2015. PMID: 26622315 Free PMC article.
-
Abnormal DNA methylation may contribute to the progression of osteosarcoma.Mol Med Rep. 2018 Jan;17(1):193-199. doi: 10.3892/mmr.2017.7869. Epub 2017 Oct 25. Mol Med Rep. 2018. PMID: 29115427 Free PMC article.
-
Identification and validation of the methylation biomarkers of non-small cell lung cancer (NSCLC).Clin Epigenetics. 2015 Jan 22;7(1):3. doi: 10.1186/s13148-014-0035-3. eCollection 2015. Clin Epigenetics. 2015. PMID: 25657825 Free PMC article.
-
MicroRNA-191 promotes osteosarcoma cells proliferation by targeting checkpoint kinase 2.Tumour Biol. 2015 Aug;36(8):6095-101. doi: 10.1007/s13277-015-3290-9. Epub 2015 Mar 14. Tumour Biol. 2015. PMID: 25773391
References
-
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. - PubMed
-
- Morgan TR, Mandayam S, Jamal MM. Alcohol and hepatocellular carcinoma. Gastroenterology. 2004;127:S87–S96. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
