Plant cap-binding complexes eukaryotic initiation factors eIF4F and eIFISO4F: molecular specificity of subunit binding
- PMID: 21965660
- PMCID: PMC3234931
- DOI: 10.1074/jbc.M111.280099
Plant cap-binding complexes eukaryotic initiation factors eIF4F and eIFISO4F: molecular specificity of subunit binding
Abstract
The initiation of translation in eukaryotes requires a suite of eIFs that include the cap-binding complex, eIF4F. eIF4F is comprised of the subunits eIF4G and eIF4E and often the helicase, eIF4A. The eIF4G subunit serves as an assembly point for other initiation factors, whereas eIF4E binds to the 7-methyl guanosine cap of mRNA. Plants have an isozyme form of eIF4F (eIFiso4F) with comparable subunits, eIFiso4E and eIFiso4G. Plant eIF4A is very loosely associated with the plant cap-binding complexes. The specificity of interaction of the individual subunits of the two complexes was previously unknown. To address this issue, mixed complexes (eIF4E-eIFiso4G or eIFiso4E-eIF4G) were expressed and purified from Escherichia coli for biochemical analysis. The activity of the mixed complexes in in vitro translation assays correlated with the large subunit of the respective correct complex. These results suggest that the eIF4G or eIFiso4G subunits influence translational efficiency more than the cap-binding subunits. The translation assays also showed varying responses of the mRNA templates to eIF4F or eIFiso4F, suggesting that some level of mRNA discrimination is possible. The dissociation constants for the correct complexes have K(D) values in the subnanomolar range, whereas the mixed complexes were found to have K(D) values in the ∼10 nm range. Displacement assays showed that the correct binding partner readily displaces the incorrect binding partner in a manner consistent with the difference in K(D) values. These results show molecular specificity for the formation of plant eIF4F and eIFiso4F complexes and suggest a role in mRNA discrimination during initiation of translation.
Figures

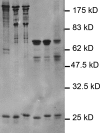
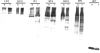
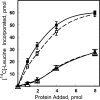
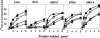


Similar articles
-
Tobacco etch virus mRNA preferentially binds wheat germ eukaryotic initiation factor (eIF) 4G rather than eIFiso4G.J Biol Chem. 2006 Nov 24;281(47):35826-34. doi: 10.1074/jbc.M605762200. Epub 2006 Sep 29. J Biol Chem. 2006. PMID: 17012235
-
The 3' cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation.RNA. 2008 Jan;14(1):134-47. doi: 10.1261/rna.777308. Epub 2007 Nov 19. RNA. 2008. PMID: 18025255 Free PMC article.
-
eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs.J Biol Chem. 2001 Oct 5;276(40):36951-60. doi: 10.1074/jbc.M103869200. Epub 2001 Aug 1. J Biol Chem. 2001. PMID: 11483601
-
eIF4F: a retrospective.J Biol Chem. 2015 Oct 2;290(40):24091-9. doi: 10.1074/jbc.R115.675280. Epub 2015 Aug 31. J Biol Chem. 2015. PMID: 26324716 Free PMC article. Review.
-
The role of the poly(A) binding protein in the assembly of the Cap-binding complex during translation initiation in plants.Translation (Austin). 2014 Oct 30;2(2):e959378. doi: 10.4161/2169074X.2014.959378. eCollection 2014 Sep 1. Translation (Austin). 2014. PMID: 26779409 Free PMC article. Review.
Cited by
-
The Triticum Mosaic Virus 5' Leader Binds to Both eIF4G and eIFiso4G for Translation.PLoS One. 2017 Jan 3;12(1):e0169602. doi: 10.1371/journal.pone.0169602. eCollection 2017. PLoS One. 2017. PMID: 28046134 Free PMC article.
-
A Dehydration-Induced Eukaryotic Translation Initiation Factor iso4G Identified in a Slow Wilting Soybean Cultivar Enhances Abiotic Stress Tolerance in Arabidopsis.Front Plant Sci. 2018 Mar 2;9:262. doi: 10.3389/fpls.2018.00262. eCollection 2018. Front Plant Sci. 2018. PMID: 29552022 Free PMC article.
-
Plant Translation Factors and Virus Resistance.Viruses. 2015 Jun 24;7(7):3392-419. doi: 10.3390/v7072778. Viruses. 2015. PMID: 26114476 Free PMC article. Review.
-
Regulation of Translation Initiation under Biotic and Abiotic Stresses.Int J Mol Sci. 2013 Feb 26;14(3):4670-83. doi: 10.3390/ijms14034670. Int J Mol Sci. 2013. PMID: 23443165 Free PMC article.
-
Recessive Resistance to Plant Viruses: Potential Resistance Genes Beyond Translation Initiation Factors.Front Microbiol. 2016 Oct 26;7:1695. doi: 10.3389/fmicb.2016.01695. eCollection 2016. Front Microbiol. 2016. PMID: 27833593 Free PMC article. Review.
References
-
- Marintchev A., Wagner G. (2004) Q. Rev. Biophys. 37, 197–284 - PubMed
-
- Pestova T. V., Lorsch J. R., Hellen C. U. (2007) in Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., Hershey J. W. eds) pp. 87–128, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
-
- Mathews M. B., Sonenberg N., Hershey J. W. (2007) in Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., Hershey J. W. eds.) pp. 1–40, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
-
- Livingstone M., Atas E., Meller A., Sonenberg N. (2010) Phys. Biol. 7, 021001 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

