Epstein-Barr virus BamHI W repeat number limits EBNA2/EBNA-LP coexpression in newly infected B cells and the efficiency of B-cell transformation: a rationale for the multiple W repeats in wild-type virus strains
- PMID: 21957300
- PMCID: PMC3209410
- DOI: 10.1128/JVI.06059-11
Epstein-Barr virus BamHI W repeat number limits EBNA2/EBNA-LP coexpression in newly infected B cells and the efficiency of B-cell transformation: a rationale for the multiple W repeats in wild-type virus strains
Abstract
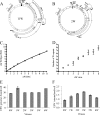
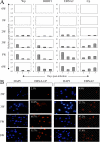
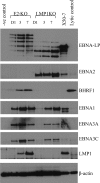
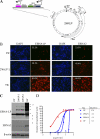
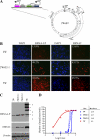
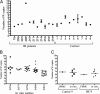
The genome of Epstein-Barr virus (EBV), a gammaherpesvirus with potent B-cell growth-transforming ability, contains multiple copies of a 3-kb BamHI W repeat sequence; each repeat carries (i) a promoter (Wp) that initiates transformation by driving EBNA-LP and EBNA2 expression and (ii) the W1W2 exons encoding the functionally active repeat domain of EBNA-LP. The W repeat copy number of a virus therefore influences two potential determinants of its transforming ability: the number of available Wp copies and the maximum size of the encoded EBNA-LP. Here, using recombinant EBVs, we show that optimal B-cell transformation requires a minimum of 5 W repeats (5W); the levels of transforming ability fall progressively with viruses carrying 4, 3, and 2 W repeats, as do the levels of Wp-initiated transcripts expressed early postinfection (p.i.), while viruses with 1 copy of the wild-type W repeat (1W) and 0W are completely nontransforming. We therefore suggest that genetic analyses of EBV transforming function should ensure that wild-type and mutant strains have equal numbers (ideally at least 5) of W copies if the analysis is not to be compromised. Attempts to enhance the transforming function of low-W-copy-number viruses, via the activity of helper EBV strains or by gene repair, suggested that the critical defect is not related to EBNA-LP size but to the failure to achieve sufficiently strong coexpression of EBNA-LP and EBNA2 early postinfection. We further show by the results of ex vivo assays that EBV strains in the blood of infected individuals typically have a mean of 5 to 8 W copies, consistent with the view that evolution has selected for viruses with an optimal transforming function.
Figures


Similar articles
-
Epstein-Barr virus nuclear antigen EBNA-LP is essential for transforming naïve B cells, and facilitates recruitment of transcription factors to the viral genome.PLoS Pathog. 2018 Feb 20;14(2):e1006890. doi: 10.1371/journal.ppat.1006890. eCollection 2018 Feb. PLoS Pathog. 2018. PMID: 29462212 Free PMC article.
-
EBNA2-deleted Epstein-Barr virus (EBV) isolate, P3HR1, causes Hodgkin-like lymphomas and diffuse large B cell lymphomas with type II and Wp-restricted latency types in humanized mice.PLoS Pathog. 2020 Jun 15;16(6):e1008590. doi: 10.1371/journal.ppat.1008590. eCollection 2020 Jun. PLoS Pathog. 2020. PMID: 32542010 Free PMC article.
-
EBNA2 and Its Coactivator EBNA-LP.Curr Top Microbiol Immunol. 2015;391:35-59. doi: 10.1007/978-3-319-22834-1_2. Curr Top Microbiol Immunol. 2015. PMID: 26428371 Review.
-
Genetic analysis of the Epstein-Barr virus-coded leader protein EBNA-LP as a co-activator of EBNA2 function.J Gen Virol. 2001 Dec;82(Pt 12):3067-3079. doi: 10.1099/0022-1317-82-12-3067. J Gen Virol. 2001. PMID: 11714985
-
Cell transformation induced by Epstein-Barr virus--living dangerously.Semin Cancer Biol. 2001 Dec;11(6):403-5. doi: 10.1006/scbi.2000.0406. Semin Cancer Biol. 2001. PMID: 11669601 Review. No abstract available.
Cited by
-
Epstein-Barr Virus-Associated Malignancies: Roles of Viral Oncoproteins in Carcinogenesis.Front Oncol. 2018 Aug 2;8:265. doi: 10.3389/fonc.2018.00265. eCollection 2018. Front Oncol. 2018. PMID: 30116721 Free PMC article. Review.
-
EBV and Apoptosis: The Viral Master Regulator of Cell Fate?Viruses. 2017 Nov 13;9(11):339. doi: 10.3390/v9110339. Viruses. 2017. PMID: 29137176 Free PMC article. Review.
-
Viral noncoding RNAs: more surprises.Genes Dev. 2015 Mar 15;29(6):567-84. doi: 10.1101/gad.259077.115. Genes Dev. 2015. PMID: 25792595 Free PMC article. Review.
-
Epstein-Barr virus protein EBNA-LP engages YY1 through leucine-rich motifs to promote naïve B cell transformation.PLoS Pathog. 2024 Jul 31;20(7):e1011950. doi: 10.1371/journal.ppat.1011950. eCollection 2024 Jul. PLoS Pathog. 2024. PMID: 39083560 Free PMC article.
-
Epstein-barr virus sequence variation-biology and disease.Pathogens. 2012 Nov 8;1(2):156-74. doi: 10.3390/pathogens1020156. Pathogens. 2012. PMID: 25436768 Free PMC article. Review.
References
-
- Allan G. J., Rowe D. T. 1989. Size and stability of the Epstein-Barr virus major internal repeat (IR-1) in Burkitt's lymphoma and lymphoblastoid cell lines. Virology 173:489–498 - PubMed
-
- Baer R., et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207–211 - PubMed
-
- Bell A., Skinner J., Kirby H., Rickinson A. 1998. Characterisation of regulatory sequences at the Epstein-Barr virus BamHI W promoter. Virology 252:149–161 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

