A functional interface at the rDNA connects rRNA synthesis, pre-rRNA processing and nucleolar surveillance in budding yeast
- PMID: 21949810
- PMCID: PMC3176313
- DOI: 10.1371/journal.pone.0024962
A functional interface at the rDNA connects rRNA synthesis, pre-rRNA processing and nucleolar surveillance in budding yeast
Abstract
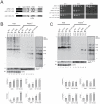
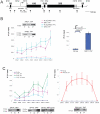
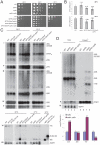
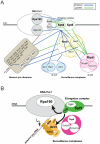
Ribogenesis is a multistep error-prone process that is actively monitored by quality control mechanisms. How ribosomal RNA synthesis, pre-rRNA processing and nucleolar surveillance are integrated is unclear. Nor is it understood how defective ribosomes are recognized. We report in budding yeast that, in vivo, the interaction between the transcription elongation factor Spt5 and Rpa190, the largest subunit of RNA polymerase (Pol) I, requires the Spt5 C-terminal region (CTR), a conserved and highly repetitive domain that is reminiscent of the RNA Pol II C-terminal domain (CTD). We show that this sequence is also required for the interaction between Spt5 and Nrd1, an RNA specific binding protein, and an exosome cofactor. Both the Spt4-Spt5, and the Nrd1-Nab3 complexes interact functionally with Rrp6, and colocalize at the rDNA. Mutations in the RNA binding domain of Nrd1, but not in its RNA Pol II CTD-interacting domain, and mutations in the RRM of Nab3 led to the accumulation of normal and aberrant polyadenylated pre-rRNAs. Altogether these results indicate that Nrd1-Nab3 contributes to recruiting the nucleolar surveillance to elongating polymerases to survey nascent rRNA transcripts.
Conflict of interest statement
Figures


Similar articles
-
The RNA polymerase II C-terminal domain-interacting domain of yeast Nrd1 contributes to the choice of termination pathway and couples to RNA processing by the nuclear exosome.J Biol Chem. 2013 Dec 20;288(51):36676-90. doi: 10.1074/jbc.M113.508267. Epub 2013 Nov 6. J Biol Chem. 2013. PMID: 24196955 Free PMC article.
-
Separable functions of the fission yeast Spt5 carboxyl-terminal domain (CTD) in capping enzyme binding and transcription elongation overlap with those of the RNA polymerase II CTD.Mol Cell Biol. 2010 May;30(10):2353-64. doi: 10.1128/MCB.00116-10. Epub 2010 Mar 15. Mol Cell Biol. 2010. PMID: 20231361 Free PMC article.
-
Yeast transcription elongation factor Spt5 associates with RNA polymerase I and RNA polymerase II directly.J Biol Chem. 2011 May 27;286(21):18825-33. doi: 10.1074/jbc.M110.202119. Epub 2011 Apr 5. J Biol Chem. 2011. PMID: 21467036 Free PMC article.
-
Beyond rRNA: nucleolar transcription generates a complex network of RNAs with multiple roles in maintaining cellular homeostasis.Genes Dev. 2022 Aug 1;36(15-16):876-886. doi: 10.1101/gad.349969.122. Genes Dev. 2022. PMID: 36207140 Free PMC article. Review.
-
The pleiotropic roles of SPT5 in transcription.Transcription. 2022 Feb-Jun;13(1-3):53-69. doi: 10.1080/21541264.2022.2103366. Epub 2022 Jul 25. Transcription. 2022. PMID: 35876486 Free PMC article. Review.
Cited by
-
Trm112 is required for Bud23-mediated methylation of the 18S rRNA at position G1575.Mol Cell Biol. 2012 Jun;32(12):2254-67. doi: 10.1128/MCB.06623-11. Epub 2012 Apr 9. Mol Cell Biol. 2012. PMID: 22493060 Free PMC article.
-
Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes.Subcell Biochem. 2013;61:211-36. doi: 10.1007/978-94-007-4525-4_10. Subcell Biochem. 2013. PMID: 23150253 Free PMC article. Review.
-
Diverse Regulators of Human Ribosome Biogenesis Discovered by Changes in Nucleolar Number.Cell Rep. 2018 Feb 13;22(7):1923-1934. doi: 10.1016/j.celrep.2018.01.056. Cell Rep. 2018. PMID: 29444442 Free PMC article.
-
Termination of Transcription of Short Noncoding RNAs by RNA Polymerase II.Annu Rev Biochem. 2015;84:381-404. doi: 10.1146/annurev-biochem-060614-034457. Epub 2015 Mar 26. Annu Rev Biochem. 2015. PMID: 25747400 Free PMC article. Review.
-
Altered rRNA processing disrupts nuclear RNA homeostasis via competition for the poly(A)-binding protein Nab2.Nucleic Acids Res. 2020 Nov 18;48(20):11675-11694. doi: 10.1093/nar/gkaa964. Nucleic Acids Res. 2020. PMID: 33137177 Free PMC article.
References
-
- Lafontaine DLJ. A ‘garbage can’ for ribosomes: how eukaryotes degrade their ribosomes? Trends Biochem Sci. 2010;35:267–277. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

