Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS
- PMID: 21944778
- PMCID: PMC3202986
- DOI: 10.1016/j.neuron.2011.09.011
Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS
Abstract
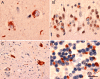
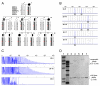
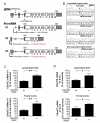
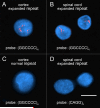
Several families have been reported with autosomal-dominant frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS), genetically linked to chromosome 9p21. Here, we report an expansion of a noncoding GGGGCC hexanucleotide repeat in the gene C9ORF72 that is strongly associated with disease in a large FTD/ALS kindred, previously reported to be conclusively linked to chromosome 9p. This same repeat expansion was identified in the majority of our families with a combined FTD/ALS phenotype and TDP-43-based pathology. Analysis of extended clinical series found the C9ORF72 repeat expansion to be the most common genetic abnormality in both familial FTD (11.7%) and familial ALS (23.5%). The repeat expansion leads to the loss of one alternatively spliced C9ORF72 transcript and to formation of nuclear RNA foci, suggesting multiple disease mechanisms. Our findings indicate that repeat expansion in C9ORF72 is a major cause of both FTD and ALS.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

Comment in
-
A hexanucleotide repeat expansion in C9ORF72 links amyotrophic lateral sclerosis and frontotemporal dementia.Nat Rev Neurol. 2011 Oct 18;7(11):595. doi: 10.1038/nrneurol.2011.162. Nat Rev Neurol. 2011. PMID: 22009283 No abstract available.
-
FTD and ALS: genetic ties that bind.Neuron. 2011 Oct 20;72(2):189-90. doi: 10.1016/j.neuron.2011.10.001. Neuron. 2011. PMID: 22017980
-
A nasty hex on chromosome 9 causes FTD/ALS.Clin Genet. 2012 Feb;81(2):126-7. doi: 10.1111/j.1399-0004.2011.01820.x. Epub 2011 Dec 28. Clin Genet. 2012. PMID: 22129088 No abstract available.
-
New gene for ALS–FTD.Mov Disord. 2012 Feb;27(2):202. doi: 10.1002/mds.24904. Mov Disord. 2012. PMID: 22423382 No abstract available.
Similar articles
-
A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD.Neuron. 2011 Oct 20;72(2):257-68. doi: 10.1016/j.neuron.2011.09.010. Epub 2011 Sep 21. Neuron. 2011. PMID: 21944779 Free PMC article.
-
C9orf72 hexanucleotide repeat expansions as the causative mutation for chromosome 9p21-linked amyotrophic lateral sclerosis and frontotemporal dementia.Arch Neurol. 2012 Sep;69(9):1159-63. doi: 10.1001/archneurol.2012.377. Arch Neurol. 2012. PMID: 22964911
-
Clinical and pathological features of familial frontotemporal dementia caused by C9ORF72 mutation on chromosome 9p.Brain. 2012 Mar;135(Pt 3):709-22. doi: 10.1093/brain/awr354. Epub 2012 Feb 17. Brain. 2012. PMID: 22344582 Free PMC article.
-
Pathogenic determinants and mechanisms of ALS/FTD linked to hexanucleotide repeat expansions in the C9orf72 gene.Neurosci Lett. 2017 Jan 1;636:16-26. doi: 10.1016/j.neulet.2016.09.007. Epub 2016 Sep 13. Neurosci Lett. 2017. PMID: 27619540 Free PMC article. Review.
-
Molecular Mechanisms of Neurodegeneration Related to C9orf72 Hexanucleotide Repeat Expansion.Behav Neurol. 2019 Jan 15;2019:2909168. doi: 10.1155/2019/2909168. eCollection 2019. Behav Neurol. 2019. PMID: 30774737 Free PMC article. Review.
Cited by
-
Plasma Small Extracellular Vesicle Cathepsin D Dysregulation in GRN/C9orf72 and Sporadic Frontotemporal Lobar Degeneration.Int J Mol Sci. 2022 Sep 14;23(18):10693. doi: 10.3390/ijms231810693. Int J Mol Sci. 2022. PMID: 36142612 Free PMC article.
-
The Repeat Expansion Diseases: The dark side of DNA repair.DNA Repair (Amst). 2015 Aug;32:96-105. doi: 10.1016/j.dnarep.2015.04.019. Epub 2015 Apr 30. DNA Repair (Amst). 2015. PMID: 26002199 Free PMC article. Review.
-
Targeting the ALS/FTD-associated A-DNA kink with anthracene-based metal complex causes DNA backbone straightening and groove contraction.Nucleic Acids Res. 2021 Sep 20;49(16):9526-9538. doi: 10.1093/nar/gkab227. Nucleic Acids Res. 2021. PMID: 33836081 Free PMC article.
-
MicroRNA signatures in genetic frontotemporal dementia and amyotrophic lateral sclerosis.Ann Clin Transl Neurol. 2022 Nov;9(11):1778-1791. doi: 10.1002/acn3.51674. Epub 2022 Oct 20. Ann Clin Transl Neurol. 2022. PMID: 36264717 Free PMC article.
-
Reduced Tau protein expression is associated with frontotemporal degeneration with progranulin mutation.Acta Neuropathol Commun. 2016 Jul 19;4(1):74. doi: 10.1186/s40478-016-0345-0. Acta Neuropathol Commun. 2016. PMID: 27435172 Free PMC article.
References
-
- Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. - PubMed
-
- Baumer D, Ansorge O, Almeida M, Talbot K. The role of RNA processing in the pathogenesis of motor neuron degeneration. Expert Rev Mol Med. 2010;12:e21. - PubMed
-
- Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG031278/AG/NIA NIH HHS/United States
- R01 AG038791/AG/NIA NIH HHS/United States
- P50NS072187/NS/NINDS NIH HHS/United States
- R01AG031278/AG/NIA NIH HHS/United States
- P01 AG019724/AG/NIA NIH HHS/United States
- P50 NS072187/NS/NINDS NIH HHS/United States
- R01AG038791/AG/NIA NIH HHS/United States
- R01 AG026251/AG/NIA NIH HHS/United States
- R01 AG026251-04/AG/NIA NIH HHS/United States
- 1RC2NS070276/NS/NINDS NIH HHS/United States
- R01 NS057567/NS/NINDS NIH HHS/United States
- P50 AG023501/AG/NIA NIH HHS/United States
- P50 AG016574/AG/NIA NIH HHS/United States
- 74580/CAPMC/ CIHR/Canada
- NS057567/NS/NINDS NIH HHS/United States
- P50AG023501/AG/NIA NIH HHS/United States
- P50 AG1657303/AG/NIA NIH HHS/United States
- 179009/CAPMC/ CIHR/Canada
- R01 NS065782-04/NS/NINDS NIH HHS/United States
- P50 AG016574-14/AG/NIA NIH HHS/United States
- RC2 NS070276/NS/NINDS NIH HHS/United States
- R01 NS065782/NS/NINDS NIH HHS/United States
- R01 AG037491/AG/NIA NIH HHS/United States
- U01 AG006786/AG/NIA NIH HHS/United States
- P01AG019724/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

