Relation between molecular shape and the morphology of self-assembling aggregates: a simulation study
- PMID: 21943424
- PMCID: PMC3177051
- DOI: 10.1016/j.bpj.2011.07.046
Relation between molecular shape and the morphology of self-assembling aggregates: a simulation study
Abstract
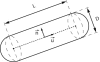
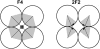
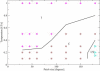
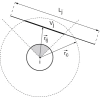
Proteins can aggregate in a wide variety of structures, both compact and extended. We present simulations of a coarse-grained anisotropic model that reproduce many of the experimentally observed aggregate structures. Conversely, all structures predicted by our model have experimental counterparts (ribbons, multistranded fibrils, and vesicles). The model we use is that of a rodlike particle with an attractive (hydrophobic) stripe on its side. Our Monte Carlo simulations show that aggregate morphologies crucially depend on two parameters. The first one is the width of the attractive stripe and the second one is a presence or absence of attractive interactions at the particle ends. These results provide us with a generic insight into the relation between the shape of protein-protein interaction potential and the morphology of protein aggregates.
Copyright © 2011 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Effects of hydrophobic interaction strength on the self-assembled structures of model peptides.Soft Matter. 2014 Jul 21;10(27):4956-65. doi: 10.1039/c4sm00378k. Epub 2014 Jun 2. Soft Matter. 2014. PMID: 24888420
-
Coarse-grained lattice model for investigating the role of cooperativity in molecular recognition.Phys Rev E Stat Nonlin Soft Matter Phys. 2007 Sep;76(3 Pt 1):031914. doi: 10.1103/PhysRevE.76.031914. Epub 2007 Sep 14. Phys Rev E Stat Nonlin Soft Matter Phys. 2007. PMID: 17930278
-
Emergent complexity from simple anisotropic building blocks: shells, tubes, and spirals.ACS Nano. 2010 Jan 26;4(1):219-28. doi: 10.1021/nn9013565. ACS Nano. 2010. PMID: 20055436
-
Modeling of Protein Structural Flexibility and Large-Scale Dynamics: Coarse-Grained Simulations and Elastic Network Models.Int J Mol Sci. 2018 Nov 6;19(11):3496. doi: 10.3390/ijms19113496. Int J Mol Sci. 2018. PMID: 30404229 Free PMC article. Review.
-
Coarse-grained representation of protein flexibility. Foundations, successes, and shortcomings.Adv Protein Chem Struct Biol. 2011;85:183-215. doi: 10.1016/B978-0-12-386485-7.00005-3. Adv Protein Chem Struct Biol. 2011. PMID: 21920324 Review.
Cited by
-
Connecting macroscopic observables and microscopic assembly events in amyloid formation using coarse grained simulations.PLoS Comput Biol. 2012;8(10):e1002692. doi: 10.1371/journal.pcbi.1002692. Epub 2012 Oct 11. PLoS Comput Biol. 2012. PMID: 23071427 Free PMC article.
-
Computational Models for the Study of Protein Aggregation.Methods Mol Biol. 2022;2340:51-78. doi: 10.1007/978-1-0716-1546-1_4. Methods Mol Biol. 2022. PMID: 35167070
-
Enhanced translocation of amphiphilic peptides across membranes by transmembrane proteins.Biophys J. 2021 Jun 1;120(11):2296-2305. doi: 10.1016/j.bpj.2021.04.005. Epub 2021 Apr 20. Biophys J. 2021. PMID: 33864790 Free PMC article.
-
Role of non-specific interactions in the phase-separation and maturation of macromolecules.PLoS Comput Biol. 2022 May 9;18(5):e1010067. doi: 10.1371/journal.pcbi.1010067. eCollection 2022 May. PLoS Comput Biol. 2022. PMID: 35533203 Free PMC article.
-
Beyond icosahedral symmetry in packings of proteins in spherical shells.Proc Natl Acad Sci U S A. 2017 Aug 22;114(34):9014-9019. doi: 10.1073/pnas.1706825114. Epub 2017 Aug 8. Proc Natl Acad Sci U S A. 2017. PMID: 28790186 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

