Maf1 protein, repressor of RNA polymerase III, indirectly affects tRNA processing
- PMID: 21940626
- PMCID: PMC3234771
- DOI: 10.1074/jbc.M111.253310
Maf1 protein, repressor of RNA polymerase III, indirectly affects tRNA processing
Abstract
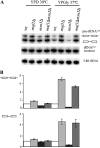
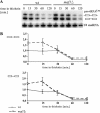
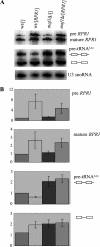
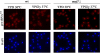
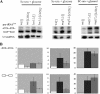
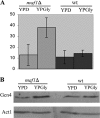
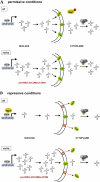
Maf1 is negative regulator of RNA polymerase III in yeast. We observed high levels of both primary transcript and end-matured, intron-containing pre-tRNAs in the maf1Δ strain. This pre-tRNA accumulation could be overcome by transcription inhibition, arguing against a direct role of Maf1 in tRNA maturation and suggesting saturation of processing machinery by the increased amounts of primary transcripts. Saturation of the tRNA exportin, Los1, is one reason why end-matured intron-containing pre-tRNAs accumulate in maf1Δ cells. However, it is likely possible that other components of the processing pathway are also limiting when tRNA transcription is increased. According to our model, Maf1-mediated transcription control and nuclear export by Los1 are two major stages of tRNA biosynthesis that are regulated by environmental conditions in a coordinated manner.
Figures

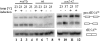

Similar articles
-
Maf1, repressor of tRNA transcription, is involved in the control of gluconeogenetic genes in Saccharomyces cerevisiae.Gene. 2013 Aug 15;526(1):16-22. doi: 10.1016/j.gene.2013.04.055. Epub 2013 May 5. Gene. 2013. PMID: 23657116
-
Maf1-mediated regulation of yeast RNA polymerase III is correlated with CCA addition at the 3' end of tRNA precursors.Gene. 2017 May 15;612:12-18. doi: 10.1016/j.gene.2016.08.033. Epub 2016 Aug 27. Gene. 2017. PMID: 27575455 Free PMC article.
-
Maf1, a general negative regulator of RNA polymerase III in yeast.Biochim Biophys Acta. 2013 Mar-Apr;1829(3-4):376-84. doi: 10.1016/j.bbagrm.2012.11.004. Epub 2012 Nov 28. Biochim Biophys Acta. 2013. PMID: 23201230 Review.
-
Regulation of RNA polymerase III transcription by Maf1 protein.Acta Biochim Pol. 2008;55(2):215-25. Epub 2008 Jun 17. Acta Biochim Pol. 2008. PMID: 18560610 Review.
-
Casein kinase II-mediated phosphorylation of general repressor Maf1 triggers RNA polymerase III activation.Proc Natl Acad Sci U S A. 2011 Mar 22;108(12):4926-31. doi: 10.1073/pnas.1010010108. Epub 2011 Mar 7. Proc Natl Acad Sci U S A. 2011. PMID: 21383183 Free PMC article.
Cited by
-
Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae.Genetics. 2013 May;194(1):43-67. doi: 10.1534/genetics.112.147470. Genetics. 2013. PMID: 23633143 Free PMC article. Review.
-
Active Center Control of Termination by RNA Polymerase III and tRNA Gene Transcription Levels In Vivo.PLoS Genet. 2016 Aug 12;12(8):e1006253. doi: 10.1371/journal.pgen.1006253. eCollection 2016 Aug. PLoS Genet. 2016. PMID: 27518095 Free PMC article.
-
Surveillance and cleavage of eukaryotic tRNAs.Int J Mol Sci. 2015 Jan 15;16(1):1873-93. doi: 10.3390/ijms16011873. Int J Mol Sci. 2015. PMID: 25599528 Free PMC article. Review.
-
Function of TFIIIC, RNA polymerase III initiation factor, in activation and repression of tRNA gene transcription.Nucleic Acids Res. 2018 Oct 12;46(18):9444-9455. doi: 10.1093/nar/gky656. Nucleic Acids Res. 2018. PMID: 30053100 Free PMC article.
-
Separate responses of karyopherins to glucose and amino acid availability regulate nucleocytoplasmic transport.Mol Biol Cell. 2014 Sep 15;25(18):2840-52. doi: 10.1091/mbc.E14-04-0948. Epub 2014 Jul 23. Mol Biol Cell. 2014. PMID: 25057022 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

