Binding site specificity and factor redundancy in activator protein-1-driven human papillomavirus chromatin-dependent transcription
- PMID: 21937452
- PMCID: PMC3220474
- DOI: 10.1074/jbc.M111.290874
Binding site specificity and factor redundancy in activator protein-1-driven human papillomavirus chromatin-dependent transcription
Abstract
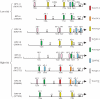
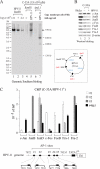
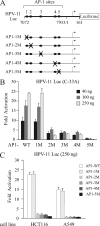
Activator protein-1 (AP-1) regulates diverse gene responses triggered by environmental cues and virus-induced cellular stress. Although many signaling events leading to AP-1 activation have been described, the fundamental features underlying binding site selection and factor recruitment of dimeric AP-1 complexes to their target genes remain mostly uncharacterized. Using recombinant full-length human AP-1 dimers formed between c-Jun and Fos family members (c-Fos, FosB, Fra-1, Fra-2) for DNA binding and transcriptional analysis, we found that each of these AP-1 complex exhibits differential activity for distinct non-consensus AP-1 sites present in human papillomavirus (HPV), and each AP-1 complex is capable of activating transcription from in vitro-reconstituted HPV chromatin in a p300- and acetyl-CoA-dependent manner. Transcription from HPV chromatin requires AP-1-dependent and contact-driven recruitment of p300. Acetylation of dimeric AP-1 complexes by p300 enhances AP-1 binding to DNA. Using a human C-33A cervical cancer-derived cell line harboring the episomal HPV type 11 genome, we illustrate binding site selectivity recognized by c-Jun, JunB, JunD, and various Fos family members in a combinatorial and unique pattern, highlighting the diversity and importance of non-canonical binding site recognition by various AP-1 family proteins.
Figures






Similar articles
-
Conversion of HPV 18 positive non-tumorigenic HeLa-fibroblast hybrids to invasive growth involves loss of TNF-alpha mediated repression of viral transcription and modification of the AP-1 transcription complex.Oncogene. 1999 May 27;18(21):3187-98. doi: 10.1038/sj.onc.1202765. Oncogene. 1999. PMID: 10359524
-
Constitutive activation of transcription factor AP-1 in cervical cancer and suppression of human papillomavirus (HPV) transcription and AP-1 activity in HeLa cells by curcumin.Int J Cancer. 2005 Mar 1;113(6):951-60. doi: 10.1002/ijc.20668. Int J Cancer. 2005. PMID: 15514944
-
Expression and distribution of AP-1 transcription factors in the porcine ovary.Biol Reprod. 2003 Jul;69(1):64-74. doi: 10.1095/biolreprod.102.013995. Epub 2003 Feb 19. Biol Reprod. 2003. PMID: 12606371
-
Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity.Oncogene. 2001 Apr 30;20(19):2438-52. doi: 10.1038/sj.onc.1204385. Oncogene. 2001. PMID: 11402339 Review.
-
The potential of activator protein 1 (AP-1) in cancer targeted therapy.Front Immunol. 2023 Jul 6;14:1224892. doi: 10.3389/fimmu.2023.1224892. eCollection 2023. Front Immunol. 2023. PMID: 37483616 Free PMC article. Review.
Cited by
-
Transregulation of microRNA miR-21 promoter by AP-1 transcription factor in cervical cancer cells.Cancer Cell Int. 2019 Aug 15;19:214. doi: 10.1186/s12935-019-0931-x. eCollection 2019. Cancer Cell Int. 2019. PMID: 31427899 Free PMC article.
-
The antiviral effects of a MEK1/2 inhibitor promote tumor regression in a preclinical model of human papillomavirus infection-induced tumorigenesis.Antiviral Res. 2023 Aug;216:105667. doi: 10.1016/j.antiviral.2023.105667. Epub 2023 Jul 8. Antiviral Res. 2023. PMID: 37429527 Free PMC article.
-
Acetylation of conserved lysines in bovine papillomavirus E2 by p300.J Virol. 2013 Feb;87(3):1497-507. doi: 10.1128/JVI.02771-12. Epub 2012 Nov 14. J Virol. 2013. PMID: 23152516 Free PMC article.
-
Papillomavirus-Associated Tumor Formation Critically Depends on c-Fos Expression Induced by Viral Protein E2 and Bromodomain Protein Brd4.PLoS Pathog. 2016 Jan 4;12(1):e1005366. doi: 10.1371/journal.ppat.1005366. eCollection 2016 Jan. PLoS Pathog. 2016. PMID: 26727473 Free PMC article.
-
A structure-based Multiple-Instance Learning approach to predicting in vitro transcription factor-DNA interaction.BMC Genomics. 2015;16 Suppl 4(Suppl 4):S3. doi: 10.1186/1471-2164-16-S4-S3. Epub 2015 Apr 21. BMC Genomics. 2015. PMID: 25917392 Free PMC article.
References
-
- Lee W., Mitchell P., Tjian R. (1987) Cell 49, 741–752 - PubMed
-
- Angel P., Karin M. (1991) Biochim. Biophys. Acta 1072, 129–157 - PubMed
-
- Jochum W., Passegué E., Wagner E. F. (2001) Oncogene 20, 2401–2412 - PubMed
-
- Mechta-Grigoriou F., Gerald D., Yaniv M. (2001) Oncogene 20, 2378–2389 - PubMed
-
- Shaulian E., Karin M. (2002) Nat. Cell Biol. 4, E131–E136 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

