Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice
- PMID: 21933985
- PMCID: PMC3198091
- DOI: 10.2337/db11-0227
Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice
Erratum in
- Diabetes. 2011 Dec;60(12):3307. Muccioli, Giulio M [corrected to Muccioli, Giulio G]
Abstract
Objective: To investigate deep and comprehensive analysis of gut microbial communities and biological parameters after prebiotic administration in obese and diabetic mice.
Research design and methods: Genetic (ob/ob) or diet-induced obese and diabetic mice were chronically fed with prebiotic-enriched diet or with a control diet. Extensive gut microbiota analyses, including quantitative PCR, pyrosequencing of the 16S rRNA, and phylogenetic microarrays, were performed in ob/ob mice. The impact of gut microbiota modulation on leptin sensitivity was investigated in diet-induced leptin-resistant mice. Metabolic parameters, gene expression, glucose homeostasis, and enteroendocrine-related L-cell function were documented in both models.
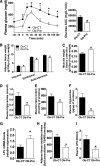
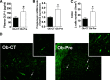
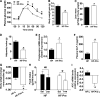
Results: In ob/ob mice, prebiotic feeding decreased Firmicutes and increased Bacteroidetes phyla, but also changed 102 distinct taxa, 16 of which displayed a >10-fold change in abundance. In addition, prebiotics improved glucose tolerance, increased L-cell number and associated parameters (intestinal proglucagon mRNA expression and plasma glucagon-like peptide-1 levels), and reduced fat-mass development, oxidative stress, and low-grade inflammation. In high fat-fed mice, prebiotic treatment improved leptin sensitivity as well as metabolic parameters.
Conclusions: We conclude that specific gut microbiota modulation improves glucose homeostasis, leptin sensitivity, and target enteroendocrine cell activity in obese and diabetic mice. By profiling the gut microbiota, we identified a catalog of putative bacterial targets that may affect host metabolism in obesity and diabetes.
Figures


Similar articles
-
Prebiotic fibres dose-dependently increase satiety hormones and alter Bacteroidetes and Firmicutes in lean and obese JCR:LA-cp rats.Br J Nutr. 2012 Feb;107(4):601-13. doi: 10.1017/S0007114511003163. Epub 2011 Jul 18. Br J Nutr. 2012. PMID: 21767445 Free PMC article.
-
Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability.Gut. 2009 Aug;58(8):1091-103. doi: 10.1136/gut.2008.165886. Epub 2009 Feb 24. Gut. 2009. PMID: 19240062 Free PMC article.
-
Inulin Can Alleviate Metabolism Disorders in ob/ob Mice by Partially Restoring Leptin-related Pathways Mediated by Gut Microbiota.Genomics Proteomics Bioinformatics. 2019 Feb;17(1):64-75. doi: 10.1016/j.gpb.2019.03.001. Epub 2019 Apr 23. Genomics Proteomics Bioinformatics. 2019. PMID: 31026583 Free PMC article.
-
Influence of diet on gut microbiota, inflammation and type 2 diabetes mellitus. First experience with macrobiotic Ma-Pi 2 diet.Diabetes Metab Res Rev. 2014 Mar;30 Suppl 1:48-54. doi: 10.1002/dmrr.2518. Diabetes Metab Res Rev. 2014. PMID: 24532292 Review.
-
Gut decontamination with norfloxacin and ampicillin enhances insulin sensitivity in mice.Nestle Nutr Workshop Ser Pediatr Program. 2008;62:127-37; discussion 137-40. doi: 10.1159/000146256. Nestle Nutr Workshop Ser Pediatr Program. 2008. PMID: 18626197 Review.
Cited by
-
Tetrahydro iso-alpha acids from hops improve glucose homeostasis and reduce body weight gain and metabolic endotoxemia in high-fat diet-fed mice.PLoS One. 2012;7(3):e33858. doi: 10.1371/journal.pone.0033858. Epub 2012 Mar 28. PLoS One. 2012. PMID: 22470484 Free PMC article.
-
Global trends in Akkermansia muciniphila research: A bibliometric visualization.Front Microbiol. 2022 Nov 10;13:1037708. doi: 10.3389/fmicb.2022.1037708. eCollection 2022. Front Microbiol. 2022. PMID: 36439840 Free PMC article.
-
Association between seaweed intake and risk of type 2 diabetes mellitus: a prospective cohort study.Br J Nutr. 2024 Apr 14;131(7):1259-1267. doi: 10.1017/S0007114523002751. Epub 2023 Nov 28. Br J Nutr. 2024. PMID: 38012847 Free PMC article.
-
Phylum level change in the cecal and fecal gut communities of rats fed diets containing different fermentable substrates supports a role for nitrogen as a factor contributing to community structure.Nutrients. 2015 May 6;7(5):3279-99. doi: 10.3390/nu7053279. Nutrients. 2015. PMID: 25954902 Free PMC article.
-
Talking microbes: When gut bacteria interact with diet and host organs.Mol Nutr Food Res. 2016 Jan;60(1):58-66. doi: 10.1002/mnfr.201500406. Epub 2015 Aug 26. Mol Nutr Food Res. 2016. PMID: 26178924 Free PMC article. Review.
References
-
- Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol 2010;72:219–246 - PubMed
-
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature 2006;444:1022–1023 - PubMed
-
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006;444:1027–1031 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

