Epigenetic regulation by RARα maintains ligand-independent transcriptional activity
- PMID: 21911359
- PMCID: PMC3245912
- DOI: 10.1093/nar/gkr637
Epigenetic regulation by RARα maintains ligand-independent transcriptional activity
Abstract
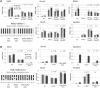
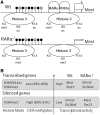
Retinoic acid receptors (RARs) α, β and γ are key regulators of embryonic development. Hematopoietic differentiation is regulated by RARα, and several types of leukemia show aberrant RARα activity. Through microarray expression analysis, we identified transcripts differentially expressed between F9 wild-type (Wt) and RARα knockout cells cultured in the absence or presence of the RAR-specific ligand all trans retinoic acid (RA). We validated the decreased Mest, Tex13, Gab1, Bcl11a, Tcfap2a and HMGcs1 transcript levels, and increased Slc38a4, Stmn2, RpL39l, Ref2L, Mobp and Rlf1 transcript levels in the RARa knockout cells. The decreased Mest and Tex13 transcript levels were associated with increased promoter CpG-island methylation and increased repressive histone modifications (H3K9me3) in RARα knockout cells. Increased Slc38a4 and Stmn2 transcript levels were associated with decreased promoter CpG-island methylation and increased permissive histone modifications (H3K9/K14ac, H3K4me3) in RARα knockout cells. We demonstrated specific association of RARα and RXRα with the Mest promoter. Importantly, stable expression of a dominant negative, oncogenic PML-RARα fusion protein in F9 Wt cells recapitulated the decreased Mest transcript levels observed in RARα knockout cells. We propose that RARα plays an important role in cellular memory and imprinting by regulating the CpG methylation status of specific promoter regions.
Figures



Similar articles
-
Differential changes of retinoid-X-receptor (RXR alpha) and its RAR alpha and PML-RAR alpha partners induced by retinoic acid and cAMP distinguish maturation sensitive and resistant t(15;17) promyelocytic leukemia NB4 cells.Oncogene. 1996 Jun 6;12(11):2443-50. Oncogene. 1996. PMID: 8649786
-
Retinoic acid receptor gamma activates receptor tyrosine kinase Tie1 gene transcription through transcription factor GATA4 in F9 stem cells.Exp Hematol. 2008 May;36(5):624-41. doi: 10.1016/j.exphem.2007.12.016. Exp Hematol. 2008. PMID: 18439490
-
Characterization of the chimeric retinoic acid receptor RARalpha/VDR.Leukemia. 1998 Apr;12(4):554-62. doi: 10.1038/sj.leu.2400937. Leukemia. 1998. PMID: 9557614
-
Interactions of STAT5b-RARalpha, a novel acute promyelocytic leukemia fusion protein, with retinoic acid receptor and STAT3 signaling pathways.Blood. 2002 Apr 15;99(8):2637-46. doi: 10.1182/blood.v99.8.2637. Blood. 2002. PMID: 11929748
-
F9 embryocarcinoma cells: a cell autonomous model to study the functional selectivity of RARs and RXRs in retinoid signaling.Histol Histopathol. 2001 Jul;16(3):909-22. doi: 10.14670/HH-16.909. Histol Histopathol. 2001. PMID: 11510982 Review.
Cited by
-
miR-1 as a Key Epigenetic Regulator in Early Differentiation of Cardiac Sinoatrial Region.Int J Mol Sci. 2024 Jun 15;25(12):6608. doi: 10.3390/ijms25126608. Int J Mol Sci. 2024. PMID: 38928314 Free PMC article.
-
Role of NADH Dehydrogenase (Ubiquinone) 1 Alpha Subcomplex 4-Like 2 in Clear Cell Renal Cell Carcinoma.Clin Cancer Res. 2016 Jun 1;22(11):2791-801. doi: 10.1158/1078-0432.CCR-15-1511. Epub 2016 Jan 18. Clin Cancer Res. 2016. PMID: 26783287 Free PMC article.
-
RARγ is essential for retinoic acid induced chromatin remodeling and transcriptional activation in embryonic stem cells.J Cell Sci. 2013 Feb 15;126(Pt 4):999-1008. doi: 10.1242/jcs.119701. Epub 2012 Dec 21. J Cell Sci. 2013. PMID: 23264745 Free PMC article.
-
A Gene Implicated in Activation of Retinoic Acid Receptor Targets Is a Novel Renal Agenesis Gene in Humans.Genetics. 2017 Sep;207(1):215-228. doi: 10.1534/genetics.117.1125. Epub 2017 Jul 24. Genetics. 2017. PMID: 28739660 Free PMC article.
-
An alternative retinoic acid-responsive Stra6 promoter regulated in response to retinol deficiency.J Biol Chem. 2015 Feb 13;290(7):4356-66. doi: 10.1074/jbc.M114.613968. Epub 2014 Dec 28. J Biol Chem. 2015. PMID: 25544292 Free PMC article.
References
-
- Collins SJ. The role of retinoids and retinoic acid receptors in normal hematopoiesis. Leukemia. 2002;16:1896–1905. - PubMed
-
- de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. - PubMed
-
- Chen Z, Guidez F, Rousselot P, Agadir A, Chen SJ, Wang ZY, Degos L, Zelent A, Waxman S, Chomienne C. PLZF-RAR alpha fusion proteins generated from the variant t(11;17)(q23;q21) translocation in acute promyelocytic leukemia inhibit ligand-dependent transactivation of wild-type retinoic acid receptors. Proc. Natl Acad. Sci. USA. 1994;91:1178–1182. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

