Global chromosomal structural instability in a subpopulation of starving Escherichia coli cells
- PMID: 21901104
- PMCID: PMC3161906
- DOI: 10.1371/journal.pgen.1002223
Global chromosomal structural instability in a subpopulation of starving Escherichia coli cells
Abstract
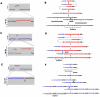
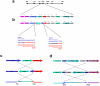
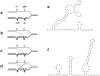
Copy-number variations (CNVs) constitute very common differences between individual humans and possibly all genomes and may therefore be important fuel for evolution, yet how they form remains elusive. In starving Escherichia coli, gene amplification is induced by stress, controlled by the general stress response. Amplification has been detected only encompassing genes that confer a growth advantage when amplified. We studied the structure of stress-induced gene amplification in starving cells in the Lac assay in Escherichia coli by array comparative genomic hybridization (aCGH), with polymerase chain reaction (pcr) and DNA sequencing to establish the structures generated. About 10% of 300 amplified isolates carried other chromosomal structural change in addition to amplification. Most of these were inversions and duplications associated with the amplification event. This complexity supports a mechanism similar to that seen in human non-recurrent copy number variants. We interpret these complex events in terms of repeated template switching during DNA replication. Importantly, we found a significant occurrence (6 out of 300) of chromosomal structural changes that were apparently not involved in the amplification event. These secondary changes were absent from 240 samples derived from starved cells not carrying amplification, suggesting that amplification happens in a differentiated subpopulation of stressed cells licensed for global chromosomal structural change and genomic instability. These data imply that chromosomal structural changes occur in bursts or showers of instability that may have the potential to drive rapid evolution.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Gross chromosomal rearrangement mediated by DNA replication in stressed cells: evidence from Escherichia coli.Ann N Y Acad Sci. 2012 Sep;1267(1):103-9. doi: 10.1111/j.1749-6632.2012.06587.x. Ann N Y Acad Sci. 2012. PMID: 22954223 Free PMC article.
-
A common copy-number breakpoint of ERBB2 amplification in breast cancer colocalizes with a complex block of segmental duplications.Breast Cancer Res. 2012 Nov 26;14(6):R150. doi: 10.1186/bcr3362. Breast Cancer Res. 2012. PMID: 23181561 Free PMC article.
-
Mechanisms for Complex Chromosomal Insertions.PLoS Genet. 2016 Nov 23;12(11):e1006446. doi: 10.1371/journal.pgen.1006446. eCollection 2016 Nov. PLoS Genet. 2016. PMID: 27880765 Free PMC article.
-
In pursuit of a molecular mechanism for adaptive gene amplification.DNA Repair (Amst). 2002 Feb 28;1(2):111-23. doi: 10.1016/s1568-7864(01)00011-8. DNA Repair (Amst). 2002. PMID: 12509258 Review.
-
Adaptive mutation and amplification in Escherichia coli: two pathways of genome adaptation under stress.Res Microbiol. 2004 Jun;155(5):352-9. doi: 10.1016/j.resmic.2004.01.020. Res Microbiol. 2004. PMID: 15207867 Review.
Cited by
-
Stress-Induced Mutagenesis, Gambler Cells, and Stealth Targeting Antibiotic-Induced Evolution.mBio. 2022 Jun 28;13(3):e0107422. doi: 10.1128/mbio.01074-22. Epub 2022 Jun 6. mBio. 2022. PMID: 35658528 Free PMC article. Review.
-
Stress-induced mutation via DNA breaks in Escherichia coli: a molecular mechanism with implications for evolution and medicine.Bioessays. 2012 Oct;34(10):885-92. doi: 10.1002/bies.201200050. Epub 2012 Aug 22. Bioessays. 2012. PMID: 22911060 Free PMC article.
-
Translesion Polymerases Drive Microhomology-Mediated Break-Induced Replication Leading to Complex Chromosomal Rearrangements.Mol Cell. 2015 Dec 17;60(6):860-72. doi: 10.1016/j.molcel.2015.10.041. Epub 2015 Dec 6. Mol Cell. 2015. PMID: 26669261 Free PMC article.
-
Stress-Induced Mutagenesis: Implications in Cancer and Drug Resistance.Annu Rev Cancer Biol. 2017 Mar;1:119-140. doi: 10.1146/annurev-cancerbio-050216-121919. Annu Rev Cancer Biol. 2017. PMID: 29399660 Free PMC article.
-
Gross chromosomal rearrangement mediated by DNA replication in stressed cells: evidence from Escherichia coli.Ann N Y Acad Sci. 2012 Sep;1267(1):103-9. doi: 10.1111/j.1749-6632.2012.06587.x. Ann N Y Acad Sci. 2012. PMID: 22954223 Free PMC article.
References
-
- Stankiewicz P, Lupski J. Structural variation in the human genome and its role in didease. Annu Rev Med. 2010;61:437–455. - PubMed
-
- Rubin CJ, Zody MC, Eriksson J, Meadows JR, Sherwood E, et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464:587–591. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

