Delineation of a core RNA element required for Kaposi's sarcoma-associated herpesvirus ORF57 binding and activity
- PMID: 21889182
- PMCID: PMC3177971
- DOI: 10.1016/j.virol.2011.08.006
Delineation of a core RNA element required for Kaposi's sarcoma-associated herpesvirus ORF57 binding and activity
Abstract
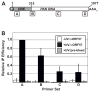
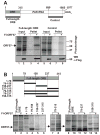
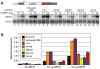
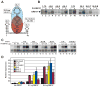
The Kaposi's sarcoma-associated herpesvirus (KSHV) ORF57 protein is an essential multifunctional regulator of gene expression. ORF57 interaction with RNA is necessary for ORF57-mediated posttranscriptional functions, but little is known about the RNA elements that drive ORF57-RNA specificity. Here, we investigate the cis-acting factors on the KSHV PAN RNA that dictate ORF57 binding and activity. We show that ORF57 binds directly to the 5' end of PAN RNA in KSHV-infected cells. Furthermore, we employ in vitro and cell-based assays to define a 30-nucleotide (nt) core ORF57-responsive element (ORE) that is necessary and sufficient for ORF57 binding and activity. Mutational analysis of the core ORE further suggests that a 9-nt sequence is a specific binding site for ORF57. These studies provide insight into ORF57 specificity determinants and lay a foundation for future analyses of cellular and viral ORF57 targets.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Kaposi's sarcoma-associated herpesvirus ORF57 protein binds and protects a nuclear noncoding RNA from cellular RNA decay pathways.PLoS Pathog. 2010 Mar 5;6(3):e1000799. doi: 10.1371/journal.ppat.1000799. PLoS Pathog. 2010. PMID: 20221435 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus ORF57 protein enhances mRNA accumulation independently of effects on nuclear RNA export.J Virol. 2007 Sep;81(18):9990-8. doi: 10.1128/JVI.00896-07. Epub 2007 Jul 3. J Virol. 2007. PMID: 17609285 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus ORF57 interacts with cellular RNA export cofactors RBM15 and OTT3 to promote expression of viral ORF59.J Virol. 2011 Feb;85(4):1528-40. doi: 10.1128/JVI.01709-10. Epub 2010 Nov 24. J Virol. 2011. PMID: 21106733 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus ORF57 in viral RNA processing.Front Biosci (Landmark Ed). 2009 Jan 1;14(4):1516-28. doi: 10.2741/3322. Front Biosci (Landmark Ed). 2009. PMID: 19273144 Free PMC article. Review.
-
New insights into the expression and functions of the Kaposi's sarcoma-associated herpesvirus long noncoding PAN RNA.Virus Res. 2016 Jan 2;212:53-63. doi: 10.1016/j.virusres.2015.06.012. Epub 2015 Jun 21. Virus Res. 2016. PMID: 26103097 Free PMC article. Review.
Cited by
-
Does the Zinc Finger Antiviral Protein (ZAP) Shape the Evolution of Herpesvirus Genomes?Viruses. 2021 Sep 17;13(9):1857. doi: 10.3390/v13091857. Viruses. 2021. PMID: 34578438 Free PMC article. Review.
-
Molecular Biology of KSHV in Relation to HIV/AIDS-Associated Oncogenesis.Cancer Treat Res. 2019;177:23-62. doi: 10.1007/978-3-030-03502-0_2. Cancer Treat Res. 2019. PMID: 30523620 Free PMC article.
-
CLIP-seq to Identify KSHV ORF57-Binding RNA in Host B Cells.Curr Protoc Microbiol. 2016 May 6;41:1E.11.1-1E.11.18. doi: 10.1002/cpmc.3. Curr Protoc Microbiol. 2016. PMID: 27153386 Free PMC article.
-
KSHV Rta Promoter Specification and Viral Reactivation.Front Microbiol. 2012 Feb 14;3:30. doi: 10.3389/fmicb.2012.00030. eCollection 2012. Front Microbiol. 2012. PMID: 22347875 Free PMC article.
-
The Kaposi's Sarcoma-Associated Herpesvirus ORF57 Protein and Its Multiple Roles in mRNA Biogenesis.Front Microbiol. 2012 Feb 20;3:59. doi: 10.3389/fmicb.2012.00059. eCollection 2012. Front Microbiol. 2012. PMID: 22363332 Free PMC article.
References
-
- Boyne JR, Whitehouse A. gamma-2 Herpes virus post-transcriptional gene regulation. Clin Microbiol Infect. 2006;12:110–117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

