PI3K/AKT signaling determines a dynamic switch between distinct KSRP functions favoring skeletal myogenesis
- PMID: 21886180
- PMCID: PMC3278731
- DOI: 10.1038/cdd.2011.117
PI3K/AKT signaling determines a dynamic switch between distinct KSRP functions favoring skeletal myogenesis
Abstract
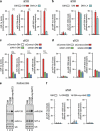
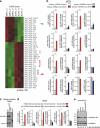
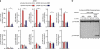
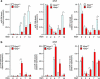
Skeletal myogenesis is orchestrated by distinct regulatory signaling pathways, including PI3K/AKT, that ultimately control muscle gene expression. Recently discovered myogenic micro-RNAs (miRNAs) are deeply implicated in muscle biology. Processing of miRNAs from their primary transcripts is emerging as a major step in the control of miRNA levels and might be well suited to be regulated by extracellular signals. Here we report that the RNA binding protein KSRP is required for the correct processing of primary myogenic miRNAs upon PI3K/AKT activation in myoblasts C2C12 and in the course of injury-induced muscle regeneration, as revealed by Ksrp knock-out mice analysis. PI3K/AKT activation regulates in opposite ways two distinct KSRP functions inhibiting its ability to promote decay of myogenin mRNA and activating its ability to favor maturation of myogenic miRNAs. This dynamic regulatory switch eventually contributes to the activation of the myogenic program.
Figures
Similar articles
-
Identification of a set of KSRP target transcripts upregulated by PI3K-AKT signaling.BMC Mol Biol. 2007 Apr 16;8:28. doi: 10.1186/1471-2199-8-28. BMC Mol Biol. 2007. PMID: 17437629 Free PMC article.
-
The RNA-binding protein KSRP promotes decay of beta-catenin mRNA and is inactivated by PI3K-AKT signaling.PLoS Biol. 2006 Dec;5(1):e5. doi: 10.1371/journal.pbio.0050005. PLoS Biol. 2006. Retraction in: PLoS Biol. 2015 Nov 16;13(11):e1002314. doi: 10.1371/journal.pbio.1002314. PMID: 17177604 Free PMC article. Retracted.
-
Trpc1 ion channel modulates phosphatidylinositol 3-kinase/Akt pathway during myoblast differentiation and muscle regeneration.J Biol Chem. 2012 Apr 27;287(18):14524-34. doi: 10.1074/jbc.M112.341784. Epub 2012 Mar 6. J Biol Chem. 2012. PMID: 22399301 Free PMC article.
-
Postnatal skeletal muscle myogenesis governed by signal transduction networks: MAPKs and PI3K-Akt control multiple steps.Biochem Biophys Res Commun. 2023 Nov 19;682:223-243. doi: 10.1016/j.bbrc.2023.09.048. Epub 2023 Sep 27. Biochem Biophys Res Commun. 2023. PMID: 37826946 Review.
-
The role of KSRP in mRNA decay and microRNA precursor maturation.Wiley Interdiscip Rev RNA. 2010 Sep-Oct;1(2):230-9. doi: 10.1002/wrna.2. Epub 2010 May 6. Wiley Interdiscip Rev RNA. 2010. PMID: 21935887 Review.
Cited by
-
Skeletal Muscle UCHL1 Negatively Regulates Muscle Development and Recovery after Muscle Injury.Int J Mol Sci. 2024 Jul 4;25(13):7330. doi: 10.3390/ijms25137330. Int J Mol Sci. 2024. PMID: 39000437 Free PMC article.
-
Sestrins regulate muscle stem cell metabolic homeostasis.Stem Cell Reports. 2021 Sep 14;16(9):2078-2088. doi: 10.1016/j.stemcr.2021.07.014. Epub 2021 Aug 12. Stem Cell Reports. 2021. PMID: 34388363 Free PMC article.
-
Noncanonical G recognition mediates KSRP regulation of let-7 biogenesis.Nat Struct Mol Biol. 2012 Dec;19(12):1282-6. doi: 10.1038/nsmb.2427. Epub 2012 Nov 11. Nat Struct Mol Biol. 2012. PMID: 23142982 Free PMC article.
-
Notch3 and Mef2c proteins are mutually antagonistic via Mkp1 protein and miR-1/206 microRNAs in differentiating myoblasts.J Biol Chem. 2012 Nov 23;287(48):40360-70. doi: 10.1074/jbc.M112.378414. Epub 2012 Oct 10. J Biol Chem. 2012. PMID: 23055528 Free PMC article.
-
MicroRNAs as Biomarkers for Early Diagnosis, Prognosis, and Therapeutic Targeting of Ovarian Cancer.J Oncol. 2021 Oct 21;2021:3408937. doi: 10.1155/2021/3408937. eCollection 2021. J Oncol. 2021. PMID: 34721577 Free PMC article. Review.
References
-
- Yun K, Wold B. Skeletal muscle determination and differentiation: story of a core regulatory network and its context. Curr Opin Cell Biol. 1996;8:877–889. - PubMed
-
- Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–238. - PubMed
-
- Tajbakhsh S. Skeletal muscle stem cells in developmental versus regenerative myogenesis. J Intern Med. 2009;266:372–389. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

