Interplay between innate immunity and negative-strand RNA viruses: towards a rational model
- PMID: 21885681
- PMCID: PMC3165544
- DOI: 10.1128/MMBR.00007-11
Interplay between innate immunity and negative-strand RNA viruses: towards a rational model
Abstract
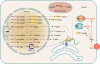
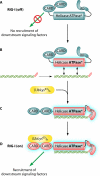
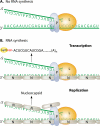
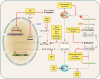
The discovery of a new class of cytosolic receptors recognizing viral RNA, called the RIG-like receptors (RLRs), has revolutionized our understanding of the interplay between viruses and host cells. A tremendous amount of work has been accumulating to decipher the RNA moieties required for an RLR agonist, the signal transduction pathway leading to activation of the innate immunity orchestrated by type I interferon (IFN), the cellular and viral regulators of this pathway, and the viral inhibitors of the innate immune response. Previous reviews have focused on the RLR signaling pathway and on the negative regulation of the interferon response by viral proteins. The focus of this review is to put this knowledge in the context of the virus replication cycle within a cell. Likewise, there has been an expansion of knowledge about the role of innate immunity in the pathophysiology of viral infection. As a consequence, some discrepancies have arisen between the current models of cell-intrinsic innate immunity and current knowledge of virus biology. This holds particularly true for the nonsegmented negative-strand viruses (Mononegavirales), which paradoxically have been largely used to build presently available models. The aim of this review is to bridge the gap between the virology and innate immunity to favor the rational building of a relevant model(s) describing the interplay between Mononegavirales and the innate immune system.
Figures

Similar articles
-
Molecular Mechanisms of Innate Immune Inhibition by Non-Segmented Negative-Sense RNA Viruses.J Mol Biol. 2016 Aug 28;428(17):3467-82. doi: 10.1016/j.jmb.2016.07.017. Epub 2016 Jul 31. J Mol Biol. 2016. PMID: 27487481 Free PMC article. Review.
-
How Dengue Virus Circumvents Innate Immunity.Front Immunol. 2018 Dec 4;9:2860. doi: 10.3389/fimmu.2018.02860. eCollection 2018. Front Immunol. 2018. PMID: 30564245 Free PMC article. Review.
-
Emerging complexity and new roles for the RIG-I-like receptors in innate antiviral immunity.Virol Sin. 2015 Jun;30(3):163-73. doi: 10.1007/s12250-015-3604-5. Epub 2015 May 20. Virol Sin. 2015. PMID: 25997992 Free PMC article. Review.
-
The RNA-binding proteins regulate innate antiviral immune signaling by modulating pattern recognition receptors.Virol J. 2024 Sep 20;21(1):225. doi: 10.1186/s12985-024-02503-x. Virol J. 2024. PMID: 39304943 Free PMC article. Review.
-
RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity.Curr Opin Virol. 2011 Sep;1(3):167-76. doi: 10.1016/j.coviro.2011.04.004. Curr Opin Virol. 2011. PMID: 21949557 Free PMC article. Review.
Cited by
-
Tick-borne Nyamanini virus replicates in the nucleus and exhibits unusual genome and matrix protein properties.J Virol. 2012 Oct;86(19):10739-47. doi: 10.1128/JVI.00571-12. Epub 2012 Jul 25. J Virol. 2012. PMID: 22837209 Free PMC article.
-
Interferon-inducible protein IFI35 negatively regulates RIG-I antiviral signaling and supports vesicular stomatitis virus replication.J Virol. 2014 Mar;88(6):3103-13. doi: 10.1128/JVI.03202-13. Epub 2013 Dec 26. J Virol. 2014. PMID: 24371060 Free PMC article.
-
New Perspectives on the Biogenesis of Viral Inclusion Bodies in Negative-Sense RNA Virus Infections.Cells. 2021 Jun 10;10(6):1460. doi: 10.3390/cells10061460. Cells. 2021. PMID: 34200781 Free PMC article. Review.
-
Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection.Mol Ther. 2013 Feb;21(2):368-79. doi: 10.1038/mt.2012.237. Epub 2012 Nov 27. Mol Ther. 2013. PMID: 23183536 Free PMC article.
-
Molecular Mechanisms of Innate Immune Inhibition by Non-Segmented Negative-Sense RNA Viruses.J Mol Biol. 2016 Aug 28;428(17):3467-82. doi: 10.1016/j.jmb.2016.07.017. Epub 2016 Jul 31. J Mol Biol. 2016. PMID: 27487481 Free PMC article. Review.
References
-
- Ahmed M., Cramer S. D., Lyles D. S. 2004. Sensitivity of prostate tumors to wild type and M protein mutant vesicular stomatitis viruses. Virology 330:34–49 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

