Cytomegalovirus antivirals and development of improved animal models
- PMID: 21883024
- PMCID: PMC4545654
- DOI: 10.1517/17425255.2011.613824
Cytomegalovirus antivirals and development of improved animal models
Abstract
Introduction: Cytomegalovirus (CMV) is a ubiquitous pathogen that establishes a lifelong asymptomatic infection in healthy individuals. Infection of immunesuppressed individuals causes serious illness. Transplant and AIDS patients are highly susceptible to CMV leading to life-threatening end-organ disease. Another vulnerable population is the developing fetus in utero, where congenital infection can result in surviving newborns with long-term developmental problems. There is no vaccine licensed for CMV and current antivirals suffer from complications associated with prolonged treatment. These include drug toxicity and emergence of resistant strains. There is an obvious need for new antivirals. Candidate intervention strategies are tested in controlled preclinical animal models but species specificity of human CMV precludes the direct study of the virus in an animal model.
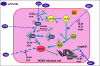
Areas covered: This review explores the current status of CMV antivirals and development of new drugs. This includes the use of animal models and the development of new improved models such as humanized animal CMV and bioluminescent imaging of virus in animals in real time.
Expert opinion: Various new CMV antivirals are in development, some with greater spectrum of activity against other viruses. Although the greatest need is in the setting of transplant patients, there remains an unmet need for a safe antiviral strategy against congenital CMV. This is especially important as an effective CMV vaccine remains an elusive goal. In this regard, greater emphasis should be placed on suitable preclinical animal models and greater collaboration between industry and academia.
Figures





Similar articles
-
Current and new cytomegalovirus antivirals and novel animal model strategies.Inflamm Allergy Drug Targets. 2010 Sep;9(4):286-99. doi: 10.2174/187152810793358822. Inflamm Allergy Drug Targets. 2010. PMID: 20860547 Review.
-
Progress toward an elusive goal: current status of cytomegalovirus vaccines.Expert Rev Vaccines. 2005 Jun;4(3):381-406. doi: 10.1586/14760584.4.3.381. Expert Rev Vaccines. 2005. PMID: 16026251 Review.
-
Nonprimate models of congenital cytomegalovirus (CMV) infection: gaining insight into pathogenesis and prevention of disease in newborns.ILAR J. 2006;47(1):65-72. doi: 10.1093/ilar.47.1.65. ILAR J. 2006. PMID: 16391432 Review.
-
Pivotal role of animal models in the development of new therapies for cytomegalovirus infections.Antiviral Res. 2006 Sep;71(2-3):164-71. doi: 10.1016/j.antiviral.2006.05.018. Epub 2006 Jun 19. Antiviral Res. 2006. PMID: 16828175 Review.
-
Clinical aspects of cytomegalovirus antiviral resistance in solid organ transplant recipients.Clin Infect Dis. 2013 Apr;56(7):1018-29. doi: 10.1093/cid/cis1035. Epub 2012 Dec 12. Clin Infect Dis. 2013. PMID: 23243176 Review.
Cited by
-
Human embryonic stem cell lines model experimental human cytomegalovirus latency.mBio. 2013 May 28;4(3):e00298-13. doi: 10.1128/mBio.00298-13. mBio. 2013. PMID: 23716573 Free PMC article.
-
A Neglected Animal Model for a Neglected Disease: Guinea Pigs and the Search for an Improved Animal Model for Human Brucellosis.Front Microbiol. 2018 Oct 31;9:2593. doi: 10.3389/fmicb.2018.02593. eCollection 2018. Front Microbiol. 2018. PMID: 30429834 Free PMC article. Review.
-
A Homolog Pentameric Complex Dictates Viral Epithelial Tropism, Pathogenicity and Congenital Infection Rate in Guinea Pig Cytomegalovirus.PLoS Pathog. 2016 Jul 7;12(7):e1005755. doi: 10.1371/journal.ppat.1005755. eCollection 2016 Jul. PLoS Pathog. 2016. PMID: 27387220 Free PMC article.
-
Viral Glycoprotein Complex Formation, Essential Function and Immunogenicity in the Guinea Pig Model for Cytomegalovirus.PLoS One. 2015 Aug 12;10(8):e0135567. doi: 10.1371/journal.pone.0135567. eCollection 2015. PLoS One. 2015. PMID: 26267274 Free PMC article.
-
Guinea pig immunoglobulin VH and VL naïve repertoire analysis.PLoS One. 2018 Dec 13;13(12):e0208977. doi: 10.1371/journal.pone.0208977. eCollection 2018. PLoS One. 2018. PMID: 30543679 Free PMC article.
References
-
- Ross SA, Boppana SB. Congenital cytomegalovirus infection: Outcome and diagnosis. Semin Pediatr Infect Dis. 2005 Jan;16(1):44–9. - PubMed
-
- Griffiths PD, Walter S. Cytomegalovirus. Curr Opin Infect Dis. 2005 Jun;18(3):241–5. - PubMed
-
- Kenneson A, Cannon MJ. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev Med Virol. 2007 Jul-Aug;17(4):253–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
