Gypsy and the birth of the SCAN domain
- PMID: 21865395
- PMCID: PMC3209298
- DOI: 10.1128/JVI.00867-11
Gypsy and the birth of the SCAN domain
Abstract
SCAN is a protein domain frequently found at the N termini of proteins encoded by mammalian tandem zinc finger (ZF) genes, whose structure is known to be similar to that of retroviral gag capsid domains and whose multimerization has been proposed as a model for retroviral assembly. We report that the SCAN domain is derived from the C-terminal portion of the gag capsid (CA) protein from the Gmr1-like family of Gypsy/Ty3-like retrotransposons. On the basis of sequence alignments and phylogenetic distributions, we show that the ancestral host SCAN domain (ESCAN for extended SCAN) was exapted from a full-length CA gene from a Gmr1-like retrotransposon at or near the root of the tetrapod animal branch. A truncated variant of ESCAN that corresponds to the annotated SCAN domain arose shortly thereafter and appears to be the only form extant in mammals. The Anolis lizard has a large number of tandem ZF genes with N-terminal ESCAN or SCAN domains. We predict DNA binding sites for all Anolis ESCAN-ZF and SCAN-ZF proteins and demonstrate several highly significant matches to Anolis Gmr1-like sequences, suggesting that at least some of these proteins target retroelements. SCAN is known to mediate protein dimerization, and the CA protein multimerizes to form the core retroviral and retrotransposon capsid structure. We speculate that the SCAN domain originally functioned to target host ZF proteins to retroelement capsids.
Figures
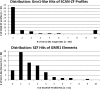
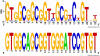
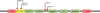
Similar articles
-
Coevolution of retroelements and tandem zinc finger genes.Genome Res. 2011 Nov;21(11):1800-12. doi: 10.1101/gr.121749.111. Epub 2011 Jul 22. Genome Res. 2011. PMID: 21784874 Free PMC article.
-
A co-opted gypsy-type LTR-retrotransposon is conserved in the genomes of humans, sheep, mice, and rats.Curr Biol. 2003 Sep 2;13(17):1518-23. doi: 10.1016/s0960-9822(03)00618-3. Curr Biol. 2003. PMID: 12956954
-
Structure of the Ty3/Gypsy retrotransposon capsid and the evolution of retroviruses.Proc Natl Acad Sci U S A. 2019 May 14;116(20):10048-10057. doi: 10.1073/pnas.1900931116. Epub 2019 Apr 29. Proc Natl Acad Sci U S A. 2019. PMID: 31036670 Free PMC article.
-
DIRS-1 and the other tyrosine recombinase retrotransposons.Cytogenet Genome Res. 2005;110(1-4):575-88. doi: 10.1159/000084991. Cytogenet Genome Res. 2005. PMID: 16093711 Review.
-
Intercellular Communication in the Nervous System Goes Viral.Trends Neurosci. 2021 Apr;44(4):248-259. doi: 10.1016/j.tins.2020.12.003. Epub 2021 Jan 21. Trends Neurosci. 2021. PMID: 33485691 Free PMC article. Review.
Cited by
-
ZBED evolution: repeated utilization of DNA transposons as regulators of diverse host functions.PLoS One. 2013;8(3):e59940. doi: 10.1371/journal.pone.0059940. Epub 2013 Mar 22. PLoS One. 2013. PMID: 23533661 Free PMC article.
-
Silencing and Transcriptional Regulation of Endogenous Retroviruses: An Overview.Viruses. 2020 Aug 13;12(8):884. doi: 10.3390/v12080884. Viruses. 2020. PMID: 32823517 Free PMC article. Review.
-
Transposable elements in human genetic disease.Nat Rev Genet. 2019 Dec;20(12):760-772. doi: 10.1038/s41576-019-0165-8. Epub 2019 Sep 12. Nat Rev Genet. 2019. PMID: 31515540 Review.
-
Extensive intron gain in the ancestor of placental mammals.Biol Direct. 2011 Nov 23;6:59. doi: 10.1186/1745-6150-6-59. Biol Direct. 2011. PMID: 22112745 Free PMC article.
-
The role of genes domesticated from LTR retrotransposons and retroviruses in mammals.Front Microbiol. 2012 Jul 27;3:262. doi: 10.3389/fmicb.2012.00262. eCollection 2012. Front Microbiol. 2012. PMID: 22866050 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

