The microbiome and rheumatoid arthritis
- PMID: 21862983
- PMCID: PMC3275101
- DOI: 10.1038/nrrheum.2011.121
The microbiome and rheumatoid arthritis
Abstract
Humans are not (and have never been) alone. From the moment we are born, millions of micro-organisms populate our bodies and coexist with us rather peacefully for the rest of our lives. This microbiome represents the totality of micro-organisms (and their genomes) that we necessarily acquire from the environment. Micro-organisms living in or on us have evolved to extract the energy they require to survive, and in exchange they support the physiological, metabolic and immune capacities that have contributed to our evolutionary success. Although currently categorized as an autoimmune disorder and regarded as a complex genetic disease, the ultimate cause of rheumatoid arthritis (RA) remains elusive. It seems that interplay between predisposing genetic factors and environmental triggers is required for disease manifestation. New insights from DNA sequence-based analyses of gut microbial communities and a renewed interest in mucosal immunology suggest that the microbiome represents an important environmental factor that can influence autoimmune disease manifestation. This Review summarizes the historical clues that suggest a possible role for the microbiota in the pathogenesis of RA, and will focus on new technologies that might provide scientific evidence to support this hypothesis.
Figures


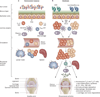

Similar articles
-
Arthritis susceptibility and the gut microbiome.FEBS Lett. 2014 Nov 17;588(22):4244-9. doi: 10.1016/j.febslet.2014.05.034. Epub 2014 May 27. FEBS Lett. 2014. PMID: 24873878 Free PMC article. Review.
-
Microbiome and probiotics: link to arthritis.Curr Opin Rheumatol. 2014 Jul;26(4):410-5. doi: 10.1097/BOR.0000000000000075. Curr Opin Rheumatol. 2014. PMID: 24841227 Review.
-
Data-driven multiple-level analysis of gut-microbiome-immune-joint interactions in rheumatoid arthritis.BMC Genomics. 2019 Feb 11;20(1):124. doi: 10.1186/s12864-019-5510-y. BMC Genomics. 2019. PMID: 30744546 Free PMC article.
-
[The role of the human microbiom in the pathogenesis of rheumatoid arthritis - a literature review].Wiad Lek. 2017;70(4):798-803. Wiad Lek. 2017. PMID: 29064808 Review. Polish.
-
Altered gut microbiota in RA: implications for treatment.Z Rheumatol. 2017 Jun;76(5):451-457. doi: 10.1007/s00393-016-0237-5. Z Rheumatol. 2017. PMID: 27909795 Review. English.
Cited by
-
Correlation of gut microbial diversity to sight-threatening diabetic retinopathy.BMC Microbiol. 2024 Sep 13;24(1):342. doi: 10.1186/s12866-024-03496-x. BMC Microbiol. 2024. PMID: 39271995 Free PMC article.
-
Alterations of mouse gut microbiome in alveolar echinococcosis.Heliyon. 2024 Jun 14;10(12):e32860. doi: 10.1016/j.heliyon.2024.e32860. eCollection 2024 Jun 30. Heliyon. 2024. PMID: 38988523 Free PMC article.
-
Gut microbiota as a sensor of autoimmune response and treatment for rheumatoid arthritis.Immunol Rev. 2024 Aug;325(1):90-106. doi: 10.1111/imr.13359. Epub 2024 Jun 12. Immunol Rev. 2024. PMID: 38867408 Review.
-
The impact of the human gut microbiome on the treatment of autoimmune disease.Immunol Rev. 2024 Aug;325(1):107-130. doi: 10.1111/imr.13358. Epub 2024 Jun 12. Immunol Rev. 2024. PMID: 38864582 Review.
-
MicroPredict: predicting species-level taxonomic abundance of whole-shotgun metagenomic data using only 16S amplicon sequencing data.Genes Genomics. 2024 Jun;46(6):701-712. doi: 10.1007/s13258-024-01514-w. Epub 2024 May 3. Genes Genomics. 2024. PMID: 38700829 Free PMC article.
References
-
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 1977;31:107–133. - PubMed
-
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host–bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. - PubMed
-
- Lederberg J. Infectious history. Science. 2000;288:287–293. - PubMed
-
- Lederberg J, McCray AT. ‘Ome sweet ‘omics—A genealogical treasury of words. Scientist. 2001;15:8–9.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

