Role of RBM25/LUC7L3 in abnormal cardiac sodium channel splicing regulation in human heart failure
- PMID: 21859973
- PMCID: PMC3172047
- DOI: 10.1161/CIRCULATIONAHA.111.044495
Role of RBM25/LUC7L3 in abnormal cardiac sodium channel splicing regulation in human heart failure
Abstract
Background: Human heart failure is associated with decreased cardiac voltage-gated Na+ channel current (encoded by SCN5A), and the changes have been implicated in the increased risk of sudden death in heart failure. Nevertheless, the mechanism of SCN5A downregulation is unclear. A number of human diseases are associated with alternative mRNA splicing, which has received comparatively little attention in the study of cardiac disease. Splicing factor expression profiles during human heart failure and a specific splicing pathway for SCN5A regulation were explored in this study.
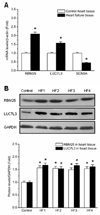
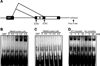
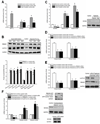
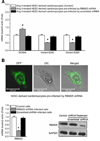
Methods and results: Gene array comparisons between normal human and heart failure tissues demonstrated that 17 splicing factors, associated with all major spliceosome components, were upregulated. Two of these splicing factors, RBM25 and LUC7L3, were elevated in human heart failure tissue and mediated truncation of SCN5A mRNA in both Jurkat cells and human embryonic stem cell-derived cardiomyocytes. RBM25/LUC7L3-mediated abnormal SCN5A mRNA splicing reduced Na+ channel current 91.1±9.3% to a range known to cause sudden death. Overexpression of either splicing factor resulted in an increase in truncated mRNA and a concomitant decrease in the full-length SCN5A transcript.
Conclusions: Of the 17 mRNA splicing factors upregulated in heart failure, RBM25 and LUC7L3 were sufficient to explain the increase in truncated forms and the reduction in full-length Na+ channel transcript. Because the reduction in channels was in the range known to be associated with sudden death, interruption of this abnormal mRNA processing may reduce arrhythmic risk in heart failure.
Figures
Similar articles
-
RBM25/LUC7L3 function in cardiac sodium channel splicing regulation of human heart failure.Trends Cardiovasc Med. 2013 Jan;23(1):5-8. doi: 10.1016/j.tcm.2012.08.003. Epub 2012 Aug 31. Trends Cardiovasc Med. 2013. PMID: 22939879 Free PMC article.
-
Human heart failure is associated with abnormal C-terminal splicing variants in the cardiac sodium channel.Circ Res. 2007 Nov 26;101(11):1146-54. doi: 10.1161/CIRCRESAHA.107.152918. Epub 2007 Sep 27. Circ Res. 2007. PMID: 17901361 Free PMC article.
-
Unfolded protein response regulates cardiac sodium current in systolic human heart failure.Circ Arrhythm Electrophysiol. 2013 Oct;6(5):1018-24. doi: 10.1161/CIRCEP.113.000274. Epub 2013 Sep 13. Circ Arrhythm Electrophysiol. 2013. PMID: 24036084 Free PMC article.
-
Structure and function of splice variants of the cardiac voltage-gated sodium channel Na(v)1.5.J Mol Cell Cardiol. 2010 Jul;49(1):16-24. doi: 10.1016/j.yjmcc.2010.04.004. Epub 2010 Apr 14. J Mol Cell Cardiol. 2010. PMID: 20398673 Review.
-
Regulation of the voltage-gated cardiac sodium channel Nav1.5 by interacting proteins.Trends Cardiovasc Med. 2005 Jan;15(1):35-40. doi: 10.1016/j.tcm.2005.01.001. Trends Cardiovasc Med. 2005. PMID: 15795161 Review.
Cited by
-
Abnormal sodium channel mRNA splicing in hypertrophic cardiomyopathy.Int J Cardiol. 2017 Dec 15;249:282-286. doi: 10.1016/j.ijcard.2017.08.071. Epub 2017 Sep 7. Int J Cardiol. 2017. PMID: 28916354 Free PMC article.
-
Melatonin to Rescue the Aged Heart: Antiarrhythmic and Antioxidant Benefits.Oxid Med Cell Longev. 2021 Mar 13;2021:8876792. doi: 10.1155/2021/8876792. eCollection 2021. Oxid Med Cell Longev. 2021. PMID: 33791076 Free PMC article. Review.
-
Differential expression of genes participating in cardiomyocyte electrophysiological remodeling via membrane ionic mechanisms and Ca2+-handling in human heart failure.Mol Cell Biochem. 2020 Jan;463(1-2):33-44. doi: 10.1007/s11010-019-03626-4. Epub 2019 Sep 13. Mol Cell Biochem. 2020. PMID: 31520233 Clinical Trial.
-
RBM25 regulates hypoxic cardiomyocyte apoptosis through CHOP-associated endoplasmic reticulum stress.Cell Stress Chaperones. 2023 Nov;28(6):861-876. doi: 10.1007/s12192-023-01380-7. Epub 2023 Sep 22. Cell Stress Chaperones. 2023. PMID: 37736860 Free PMC article.
-
Plasma cortisol-linked gene networks in hepatic and adipose tissues implicate corticosteroid-binding globulin in modulating tissue glucocorticoid action and cardiovascular risk.Front Endocrinol (Lausanne). 2023 Sep 6;14:1186252. doi: 10.3389/fendo.2023.1186252. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37745713 Free PMC article.
References
-
- Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30:13–19. - PubMed
-
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. - PubMed
-
- Novak AJ, Slager SL, Fredericksen ZS, Wang AH, Manske MM, Ziesmer S, Liebow M, Macon WR, Dillon SR, Witzig TE, Cerhan JR, Ansell SM. Genetic variation in B-cell-activating factor is associated with an increased risk of developing B-cell non-Hodgkin lymphoma. Cancer Res. 2009;69:4217–4224. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

