A copy number variation morbidity map of developmental delay
- PMID: 21841781
- PMCID: PMC3171215
- DOI: 10.1038/ng.909
A copy number variation morbidity map of developmental delay
Erratum in
- Nat Genet. 2014 Sep;46(9):1040
Abstract
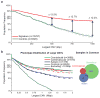
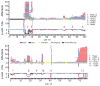
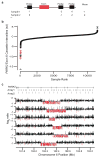
To understand the genetic heterogeneity underlying developmental delay, we compared copy number variants (CNVs) in 15,767 children with intellectual disability and various congenital defects (cases) to CNVs in 8,329 unaffected adult controls. We estimate that ∼14.2% of disease in these children is caused by CNVs >400 kb. We observed a greater enrichment of CNVs in individuals with craniofacial anomalies and cardiovascular defects compared to those with epilepsy or autism. We identified 59 pathogenic CNVs, including 14 new or previously weakly supported candidates, refined the critical interval for several genomic disorders, such as the 17q21.31 microdeletion syndrome, and identified 940 candidate dosage-sensitive genes. We also developed methods to opportunistically discover small, disruptive CNVs within the large and growing diagnostic array datasets. This evolving CNV morbidity map, combined with exome and genome sequencing, will be critical for deciphering the genetic basis of developmental delay, intellectual disability and autism spectrum disorders.
Conflict of interest statement
E.E.E. is a member of the Scientific Advisory Board of Pacific Biosciences. J.A.R., B.C.B., and L.G.S are employees of PerkinElmer.
Figures

Similar articles
-
Ohnologs are overrepresented in pathogenic copy number mutations.Proc Natl Acad Sci U S A. 2014 Jan 7;111(1):361-6. doi: 10.1073/pnas.1309324111. Epub 2013 Dec 24. Proc Natl Acad Sci U S A. 2014. PMID: 24368850 Free PMC article.
-
The clinical benefit of array-based comparative genomic hybridization for detection of copy number variants in Czech children with intellectual disability and developmental delay.BMC Med Genomics. 2019 Jul 23;12(1):111. doi: 10.1186/s12920-019-0559-7. BMC Med Genomics. 2019. PMID: 31337399 Free PMC article.
-
Genetic and phenotypic analysis of 101 patients with developmental delay or intellectual disability using whole-exome sequencing.Clin Genet. 2021 Jul;100(1):40-50. doi: 10.1111/cge.13951. Epub 2021 Mar 8. Clin Genet. 2021. PMID: 33644862
-
Interpretation of array comparative genome hybridization data: a major challenge.Cytogenet Genome Res. 2011;135(3-4):222-7. doi: 10.1159/000334066. Epub 2011 Nov 12. Cytogenet Genome Res. 2011. PMID: 22086107 Review.
-
The array CGH and its clinical applications.Drug Discov Today. 2008 Sep;13(17-18):760-70. doi: 10.1016/j.drudis.2008.06.007. Epub 2008 Jul 17. Drug Discov Today. 2008. PMID: 18617013 Review.
Cited by
-
Copy number variations and cognitive phenotypes in unselected populations.JAMA. 2015 May 26;313(20):2044-54. doi: 10.1001/jama.2015.4845. JAMA. 2015. PMID: 26010633 Free PMC article.
-
High positive predictive value of CNVs detected by clinical exome sequencing in suspected genetic diseases.J Transl Med. 2024 Jul 9;22(1):644. doi: 10.1186/s12967-024-05468-1. J Transl Med. 2024. PMID: 38982507 Free PMC article.
-
Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex.Development. 2012 May;139(9):1535-46. doi: 10.1242/dev.069963. Development. 2012. PMID: 22492350 Free PMC article. Review.
-
Discovery of variants unmasked by hemizygous deletions.Eur J Hum Genet. 2012 Jul;20(7):748-53. doi: 10.1038/ejhg.2011.263. Epub 2012 Jan 18. Eur J Hum Genet. 2012. PMID: 22258528 Free PMC article.
-
SHANK3 haploinsufficiency: a "common" but underdiagnosed highly penetrant monogenic cause of autism spectrum disorders.Mol Autism. 2013 Jun 11;4(1):17. doi: 10.1186/2040-2392-4-17. Mol Autism. 2013. PMID: 23758743 Free PMC article.
References
-
- Walsh T, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–43. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

