Interstitial contacts in an RNA-dependent RNA polymerase lattice
- PMID: 21839092
- PMCID: PMC3249232
- DOI: 10.1016/j.jmb.2011.07.053
Interstitial contacts in an RNA-dependent RNA polymerase lattice
Abstract
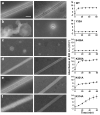
Catalytic activities can be facilitated by ordered enzymatic arrays that co-localize and orient enzymes and their substrates. The purified RNA-dependent RNA polymerase from poliovirus self-assembles to form two-dimensional lattices, possibly facilitating the assembly of viral RNA replication complexes on the cytoplasmic face of intracellular membranes. Creation of a two-dimensional lattice requires at least two different molecular contacts between polymerase molecules. One set of polymerase contacts, between the "thumb" domain of one polymerase and the back of the "palm" domain of another, has been previously defined. To identify the second interface needed for lattice formation and to test its function in viral RNA synthesis, we used a hybrid approach of electron microscopic and biochemical evaluation of both wild-type and mutant viral polymerases to evaluate computationally generated models of this second interface. A unique solution satisfied all constraints and predicted a two-dimensional structure formed from antiparallel arrays of polymerase fibers that use contacts from the flexible amino-terminal region of the protein. Enzymes that contained mutations in this newly defined interface did not form lattices and altered the structure of wild-type lattices. When reconstructed into virus, mutations that disrupt lattice assembly exhibited growth defects, synthetic lethality or both, supporting the function of the oligomeric lattice in infected cells. Understanding the structure of polymerase lattices within the multimeric RNA-dependent RNA polymerase complex should facilitate antiviral drug design and provide a precedent for other positive-strand RNA viruses.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures





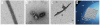
Similar articles
-
Structure-function relationships of the RNA-dependent RNA polymerase from poliovirus (3Dpol). A surface of the primary oligomerization domain functions in capsid precursor processing and VPg uridylylation.J Biol Chem. 2002 Aug 30;277(35):31551-62. doi: 10.1074/jbc.M204408200. Epub 2002 Jun 19. J Biol Chem. 2002. PMID: 12077141
-
Enzymatic and nonenzymatic functions of viral RNA-dependent RNA polymerases within oligomeric arrays.RNA. 2010 Feb;16(2):382-93. doi: 10.1261/rna.1955410. Epub 2010 Jan 5. RNA. 2010. PMID: 20051491 Free PMC article.
-
Surface for catalysis by poliovirus RNA-dependent RNA polymerase.J Mol Biol. 2013 Jul 24;425(14):2529-40. doi: 10.1016/j.jmb.2013.04.007. Epub 2013 Apr 11. J Mol Biol. 2013. PMID: 23583774 Free PMC article.
-
Picornaviral polymerase structure, function, and fidelity modulation.Virus Res. 2017 Apr 15;234:4-20. doi: 10.1016/j.virusres.2017.01.026. Epub 2017 Feb 2. Virus Res. 2017. PMID: 28163093 Free PMC article. Review.
-
Picornavirus RNA-dependent RNA polymerase.Int J Biochem Cell Biol. 2009 Mar;41(3):498-502. doi: 10.1016/j.biocel.2008.03.019. Epub 2008 Apr 7. Int J Biochem Cell Biol. 2009. PMID: 18487072 Review.
Cited by
-
Global RNA structure analysis of poliovirus identifies a conserved RNA structure involved in viral replication and infectivity.J Virol. 2013 Nov;87(21):11670-83. doi: 10.1128/JVI.01560-13. Epub 2013 Aug 21. J Virol. 2013. PMID: 23966409 Free PMC article.
-
Higher-order structures of the foot-and-mouth disease virus RNA-dependent RNA polymerase required for genome replication.Commun Biol. 2022 Jan 17;5(1):61. doi: 10.1038/s42003-021-02989-z. Commun Biol. 2022. PMID: 35039618 Free PMC article.
-
Pancreatic acinar cell-specific autophagy disruption reduces coxsackievirus replication and pathogenesis in vivo.Cell Host Microbe. 2012 Mar 15;11(3):298-305. doi: 10.1016/j.chom.2012.01.014. Cell Host Microbe. 2012. PMID: 22423969 Free PMC article.
-
A crystal structure of the dengue virus non-structural protein 5 (NS5) polymerase delineates interdomain amino acid residues that enhance its thermostability and de novo initiation activities.J Biol Chem. 2013 Oct 25;288(43):31105-14. doi: 10.1074/jbc.M113.508606. Epub 2013 Sep 11. J Biol Chem. 2013. PMID: 24025331 Free PMC article.
-
Picornaviral polymerase domain exchanges reveal a modular basis for distinct biochemical activities of viral RNA-dependent RNA polymerases.J Biol Chem. 2020 Jul 31;295(31):10624-10637. doi: 10.1074/jbc.RA120.013906. Epub 2020 Jun 3. J Biol Chem. 2020. PMID: 32493771 Free PMC article.
References
-
- Qin W, Luo H, Nomura T, Hayashi N, Yamashita T, Murakami S. Oligomeric interaction of hepatitis C virus NS5B is critical for catalytic activity of RNA-dependent RNA polymerase. J Biol Chem. 2002;277:2132–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- S10 RR025434/RR/NCRR NIH HHS/United States
- DP1 OD000827/OD/NIH HHS/United States
- T15 LM007033/LM/NLM NIH HHS/United States
- T32 AI07328/AI/NIAID NIH HHS/United States
- T32 AI007328-21/AI/NIAID NIH HHS/United States
- DP1 OD000827-05/OD/NIH HHS/United States
- R01 GM055722/GM/NIGMS NIH HHS/United States
- R01 GM055722-09/GM/NIGMS NIH HHS/United States
- GM55722/GM/NIGMS NIH HHS/United States
- S10 RR025434-01/RR/NCRR NIH HHS/United States
- R37 AI047365/AI/NIAID NIH HHS/United States
- DP1 827/DP/NCCDPHP CDC HHS/United States
- T32 AI007328/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources

