4.4 Å cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus
- PMID: 21829169
- PMCID: PMC3173789
- DOI: 10.1038/emboj.2011.261
4.4 Å cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus
Abstract
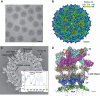
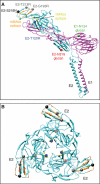
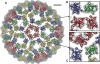
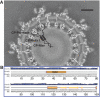
Venezuelan equine encephalitis virus (VEEV), a member of the membrane-containing Alphavirus genus, is a human and equine pathogen, and has been developed as a biological weapon. Using electron cryo-microscopy (cryo-EM), we determined the structure of an attenuated vaccine strain, TC-83, of VEEV to 4.4 Å resolution. Our density map clearly resolves regions (including E1, E2 transmembrane helices and cytoplasmic tails) that were missing in the crystal structures of domains of alphavirus subunits. These new features are implicated in the fusion, assembly and budding processes of alphaviruses. Furthermore, our map reveals the unexpected E3 protein, which is cleaved and generally thought to be absent in the mature VEEV. Our structural results suggest a mechanism for the initial stage of nucleocapsid core formation, and shed light on the virulence attenuation, host recognition and neutralizing activities of VEEV and other alphavirus pathogens.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
The SD1 Subdomain of Venezuelan Equine Encephalitis Virus Capsid Protein Plays a Critical Role in Nucleocapsid and Particle Assembly.J Virol. 2015 Dec 9;90(4):2008-20. doi: 10.1128/JVI.02680-15. Print 2016 Feb 15. J Virol. 2015. PMID: 26656680 Free PMC article.
-
Novel Insect-Specific Eilat Virus-Based Chimeric Vaccine Candidates Provide Durable, Mono- and Multivalent, Single-Dose Protection against Lethal Alphavirus Challenge.J Virol. 2018 Jan 30;92(4):e01274-17. doi: 10.1128/JVI.01274-17. Print 2018 Feb 15. J Virol. 2018. PMID: 29187545 Free PMC article.
-
A new inactivation method to facilitate cryo-EM of enveloped, RNA viruses requiring high containment: A case study using Venezuelan Equine Encephalitis Virus (VEEV).J Virol Methods. 2020 Mar;277:113792. doi: 10.1016/j.jviromet.2019.113792. Epub 2019 Nov 28. J Virol Methods. 2020. PMID: 31786314
-
Venezuelan Equine Encephalitis Virus Capsid-The Clever Caper.Viruses. 2017 Sep 29;9(10):279. doi: 10.3390/v9100279. Viruses. 2017. PMID: 28961161 Free PMC article. Review.
-
Current Understanding of the Molecular Basis of Venezuelan Equine Encephalitis Virus Pathogenesis and Vaccine Development.Viruses. 2019 Feb 18;11(2):164. doi: 10.3390/v11020164. Viruses. 2019. PMID: 30781656 Free PMC article. Review.
Cited by
-
Residue-level resolution of alphavirus envelope protein interactions in pH-dependent fusion.Proc Natl Acad Sci U S A. 2015 Feb 17;112(7):2034-9. doi: 10.1073/pnas.1414190112. Epub 2015 Feb 2. Proc Natl Acad Sci U S A. 2015. PMID: 25646410 Free PMC article.
-
Structural Insights into Alphavirus Assembly Revealed by the Cryo-EM Structure of Getah Virus.Viruses. 2022 Feb 5;14(2):327. doi: 10.3390/v14020327. Viruses. 2022. PMID: 35215918 Free PMC article.
-
Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.Cell Rep. 2018 Dec 11;25(11):3136-3147.e5. doi: 10.1016/j.celrep.2018.11.067. Cell Rep. 2018. PMID: 30540945 Free PMC article.
-
Protective antibodies against Eastern equine encephalitis virus bind to epitopes in domains A and B of the E2 glycoprotein.Nat Microbiol. 2019 Jan;4(1):187-197. doi: 10.1038/s41564-018-0286-4. Epub 2018 Nov 19. Nat Microbiol. 2019. PMID: 30455470 Free PMC article.
-
Understanding the interactability of chikungunya virus proteins via molecular recognition feature analysis.RSC Adv. 2018 Jul 31;8(48):27293-27303. doi: 10.1039/c8ra04760j. eCollection 2018 Jul 30. RSC Adv. 2018. PMID: 35539973 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

