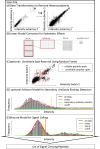
rapmad: Robust analysis of peptide microarray data
- PMID: 21816082
- PMCID: PMC3174949
- DOI: 10.1186/1471-2105-12-324
rapmad: Robust analysis of peptide microarray data
Abstract
Background: Peptide microarrays offer an enormous potential as a screening tool for peptidomics experiments and have recently seen an increased field of application ranging from immunological studies to systems biology. By allowing the parallel analysis of thousands of peptides in a single run they are suitable for high-throughput settings. Since data characteristics of peptide microarrays differ from DNA oligonucleotide microarrays, computational methods need to be tailored to these specifications to allow a robust and automated data analysis. While follow-up experiments can ensure the specificity of results, sensitivity cannot be recovered in later steps. Providing sensitivity is thus a primary goal of data analysis procedures. To this end we created rapmad (Robust Alignment of Peptide MicroArray Data), a novel computational tool implemented in R.
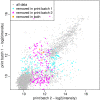
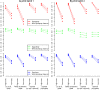
Results: We evaluated rapmad in antibody reactivity experiments for several thousand peptide spots and compared it to two existing algorithms for the analysis of peptide microarrays. rapmad displays competitive and superior behavior to existing software solutions. Particularly, it shows substantially improved sensitivity for low intensity settings without sacrificing specificity. It thereby contributes to increasing the effectiveness of high throughput screening experiments.
Conclusions: rapmad allows the robust and sensitive, automated analysis of high-throughput peptide array data. The rapmad R-package as well as the data sets are available from http://www.tron-mz.de/compmed.
Figures
Similar articles
-
Web-based design of peptide microarrays using microPepArray Pro.Methods Mol Biol. 2009;570:391-401. doi: 10.1007/978-1-60327-394-7_22. Methods Mol Biol. 2009. PMID: 19649608 Review.
-
Large-scale mining for similar protein binding pockets: with RAPMAD retrieval on the fly becomes real.J Chem Inf Model. 2015 Jan 26;55(1):165-79. doi: 10.1021/ci5005898. Epub 2014 Dec 18. J Chem Inf Model. 2015. PMID: 25474400
-
Qualitative and quantitative analysis of peptide microarray binding experiments using SVM-PEPARRAY.Methods Mol Biol. 2009;570:403-11. doi: 10.1007/978-1-60327-394-7_23. Methods Mol Biol. 2009. PMID: 19649609 Review.
-
ArrayPitope: Automated Analysis of Amino Acid Substitutions for Peptide Microarray-Based Antibody Epitope Mapping.PLoS One. 2017 Jan 17;12(1):e0168453. doi: 10.1371/journal.pone.0168453. eCollection 2017. PLoS One. 2017. PMID: 28095436 Free PMC article.
-
PMA: Protein Microarray Analyser, a user-friendly tool for data processing and normalization.BMC Res Notes. 2018 Feb 27;11(1):156. doi: 10.1186/s13104-018-3266-0. BMC Res Notes. 2018. PMID: 29482592 Free PMC article.
Cited by
-
Neutralizing Antibody Responses following Long-Term Vaccination with HIV-1 Env gp140 in Guinea Pigs.J Virol. 2018 Jun 13;92(13):e00369-18. doi: 10.1128/JVI.00369-18. Print 2018 Jul 1. J Virol. 2018. PMID: 29643249 Free PMC article.
-
Ranking Antibody Binding Epitopes and Proteins Across Samples from Whole Proteome Tiled Linear Peptides.bioRxiv [Preprint]. 2023 Apr 25:2023.04.23.536620. doi: 10.1101/2023.04.23.536620. bioRxiv. 2023. Update in: Bioinformatics. 2024 Nov 28;40(12):btae637. doi: 10.1093/bioinformatics/btae637 PMID: 37162956 Free PMC article. Updated. Preprint.
-
Identification of three immunodominant motifs with atypical isotype profile scattered over the Onchocerca volvulus proteome.PLoS Negl Trop Dis. 2017 Jan 26;11(1):e0005330. doi: 10.1371/journal.pntd.0005330. eCollection 2017 Jan. PLoS Negl Trop Dis. 2017. PMID: 28125577 Free PMC article.
-
Bayesian hierarchical modeling for subject-level response classification in peptide microarray immunoassays.Biometrics. 2016 Dec;72(4):1206-1215. doi: 10.1111/biom.12523. Epub 2016 Apr 8. Biometrics. 2016. PMID: 27061097 Free PMC article.
-
Integrating machine learning to advance epitope mapping.Front Immunol. 2024 Sep 30;15:1463931. doi: 10.3389/fimmu.2024.1463931. eCollection 2024. Front Immunol. 2024. PMID: 39403389 Free PMC article. Review.
References
-
- Lopez-Campos GH, Garcia-Albert L, Martin-Sanchez F, Garcia-Saiz A. Analysis and management of HIV peptide microarray experiments. Methods Inf Med. 2006;45:158–162. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

