Coordinated protein and DNA remodeling by human HLTF on stalled replication fork
- PMID: 21795603
- PMCID: PMC3161587
- DOI: 10.1073/pnas.1101951108
Coordinated protein and DNA remodeling by human HLTF on stalled replication fork
Abstract
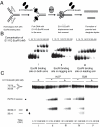
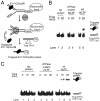
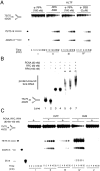
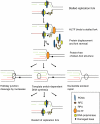
Human helicase-like transcription factor (HLTF) exhibits ubiquitin ligase activity for proliferating cell nuclear antigen (PCNA) polyubiquitylation as well as double-stranded DNA translocase activity for remodeling stalled replication fork by fork reversal, which can support damage bypass by template switching. However, a stalled replication fork is surrounded by various DNA-binding proteins which can inhibit the access of damage bypass players, and it is unknown how these proteins become displaced. Here we reveal that HLTF has an ATP hydrolysis-dependent protein remodeling activity, by which it can remove proteins bound to the replication fork. Moreover, we demonstrate that HLTF can displace a broad spectrum of proteins such as replication protein A (RPA), PCNA, and replication factor C (RFC), thereby providing the first example for a protein clearing activity at the stalled replication fork. Our findings clarify how remodeling of a stalled replication fork can occur if it is engaged in interactions with masses of proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Comment in
-
Clearance of roadblocks in replication fork restart.Proc Natl Acad Sci U S A. 2011 Aug 23;108(34):13881-2. doi: 10.1073/pnas.1110698108. Epub 2011 Aug 8. Proc Natl Acad Sci U S A. 2011. PMID: 21825169 Free PMC article. No abstract available.
Similar articles
-
Time for remodeling: SNF2-family DNA translocases in replication fork metabolism and human disease.DNA Repair (Amst). 2020 Nov;95:102943. doi: 10.1016/j.dnarep.2020.102943. Epub 2020 Aug 15. DNA Repair (Amst). 2020. PMID: 32971328 Free PMC article. Review.
-
Helicase-Like Transcription Factor HLTF and E3 Ubiquitin Ligase SHPRH Confer DNA Damage Tolerance through Direct Interactions with Proliferating Cell Nuclear Antigen (PCNA).Int J Mol Sci. 2020 Jan 21;21(3):693. doi: 10.3390/ijms21030693. Int J Mol Sci. 2020. PMID: 31973093 Free PMC article.
-
Regulation of HLTF-mediated PCNA polyubiquitination by RFC and PCNA monoubiquitination levels determines choice of damage tolerance pathway.Nucleic Acids Res. 2018 Nov 30;46(21):11340-11356. doi: 10.1093/nar/gky943. Nucleic Acids Res. 2018. PMID: 30335157 Free PMC article.
-
Human HLTF mediates postreplication repair by its HIRAN domain-dependent replication fork remodelling.Nucleic Acids Res. 2015 Dec 2;43(21):10277-91. doi: 10.1093/nar/gkv896. Epub 2015 Sep 8. Nucleic Acids Res. 2015. PMID: 26350214 Free PMC article.
-
Structure of the HLTF HIRAN domain and its functional implications in regression of a stalled replication fork.Acta Crystallogr D Struct Biol. 2020 Aug 1;76(Pt 8):729-735. doi: 10.1107/S2059798320008074. Epub 2020 Jul 27. Acta Crystallogr D Struct Biol. 2020. PMID: 32744255 Review.
Cited by
-
Preventing replication fork collapse to maintain genome integrity.DNA Repair (Amst). 2015 Aug;32:149-157. doi: 10.1016/j.dnarep.2015.04.026. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 25957489 Free PMC article. Review.
-
Time for remodeling: SNF2-family DNA translocases in replication fork metabolism and human disease.DNA Repair (Amst). 2020 Nov;95:102943. doi: 10.1016/j.dnarep.2020.102943. Epub 2020 Aug 15. DNA Repair (Amst). 2020. PMID: 32971328 Free PMC article. Review.
-
WRNIP1 Protects Reversed DNA Replication Forks from SLX4-Dependent Nucleolytic Cleavage.iScience. 2019 Nov 22;21:31-41. doi: 10.1016/j.isci.2019.10.010. Epub 2019 Oct 8. iScience. 2019. PMID: 31654852 Free PMC article.
-
Strand annealing and motor driven activities of SMARCAL1 and ZRANB3 are stimulated by RAD51 and the paralog complex.Nucleic Acids Res. 2022 Aug 12;50(14):8008-8022. doi: 10.1093/nar/gkac583. Nucleic Acids Res. 2022. PMID: 35801922 Free PMC article.
-
Loss of HLTF function promotes intestinal carcinogenesis.Mol Cancer. 2012 Mar 27;11:18. doi: 10.1186/1476-4598-11-18. Mol Cancer. 2012. PMID: 22452792 Free PMC article.
References
-
- Boulton SJ. Helicase and translocases required for the maintenance of genome stability. DNA Repair (Amst) 2010;9(3):201. - PubMed
-
- Boulton SJ. DNA repair: Decision at the break point. Nature. 2010;465:301–302. - PubMed
-
- Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

