Access to DNA establishes a secondary target site bias for the yeast retrotransposon Ty5
- PMID: 21788500
- PMCID: PMC3251128
- DOI: 10.1073/pnas.1103665108
Access to DNA establishes a secondary target site bias for the yeast retrotransposon Ty5
Abstract
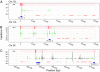
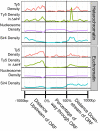
Integration sites for many retrotransposons and retroviruses are determined by interactions between retroelement-encoded integrases and specific DNA-bound proteins. The Saccharomyces retrotransposon Ty5 preferentially integrates into heterochromatin because of interactions between Ty5 integrase and the heterochromatin protein silent information regulator 4. We mapped over 14,000 Ty5 insertions onto the S. cerevisiae genome, 76% of which occurred in heterochromatin, which is consistent with the known target site bias of Ty5. Using logistic regression, associations were assessed between Ty5 insertions and various chromosomal features such as genome-wide distributions of nucleosomes and histone modifications. Sites of Ty5 insertion, regardless of whether they occurred in heterochromatin or euchromatin, were strongly associated with DNase hypersensitive, nucleosome-free regions flanking genes. Our data support a model wherein silent information regulator 4 tethers the Ty5 integration machinery to domains of heterochromatin, and then, specific target sites are selected based on DNA access, resulting in a secondary target site bias. For insertions in euchromatin, DNA access is the primary determinant of target site choice. One consequence of the secondary target site bias of Ty5 is that insertions in coding sequences occur infrequently, which may preserve genome integrity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Targeting integration of the Saccharomyces Ty5 retrotransposon.Methods Mol Biol. 2008;435:153-63. doi: 10.1007/978-1-59745-232-8_11. Methods Mol Biol. 2008. PMID: 18370074
-
Phosphorylation regulates integration of the yeast Ty5 retrotransposon into heterochromatin.Mol Cell. 2007 Jul 20;27(2):289-299. doi: 10.1016/j.molcel.2007.06.010. Mol Cell. 2007. PMID: 17643377
-
The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci.Genes Dev. 1996 Mar 1;10(5):634-45. doi: 10.1101/gad.10.5.634. Genes Dev. 1996. PMID: 8598292
-
Light and shadow on the mechanisms of integration site selection in yeast Ty retrotransposon families.Curr Genet. 2021 Jun;67(3):347-357. doi: 10.1007/s00294-021-01154-7. Epub 2021 Feb 15. Curr Genet. 2021. PMID: 33590295 Review.
-
Stress management: how cells take control of their transposons.Mol Cell. 2007 Jul 20;27(2):180-181. doi: 10.1016/j.molcel.2007.07.004. Mol Cell. 2007. PMID: 17643368 Review.
Cited by
-
Disentangling the determinants of transposable elements dynamics in vertebrate genomes using empirical evidences and simulations.PLoS Genet. 2020 Oct 5;16(10):e1009082. doi: 10.1371/journal.pgen.1009082. eCollection 2020 Oct. PLoS Genet. 2020. PMID: 33017388 Free PMC article.
-
A nucleosomal surface defines an integration hotspot for the Saccharomyces cerevisiae Ty1 retrotransposon.Genome Res. 2012 Apr;22(4):704-13. doi: 10.1101/gr.129585.111. Epub 2012 Jan 4. Genome Res. 2012. PMID: 22219511 Free PMC article.
-
Evolutionary genomics of transposable elements in Saccharomyces cerevisiae.PLoS One. 2012;7(11):e50978. doi: 10.1371/journal.pone.0050978. Epub 2012 Nov 30. PLoS One. 2012. PMID: 23226439 Free PMC article.
-
Two large-scale analyses of Ty1 LTR-retrotransposon de novo insertion events indicate that Ty1 targets nucleosomal DNA near the H2A/H2B interface.Mob DNA. 2012 Dec 17;3(1):22. doi: 10.1186/1759-8753-3-22. Mob DNA. 2012. PMID: 23244340 Free PMC article.
-
Diverse transposable element landscapes in pathogenic and nonpathogenic yeast models: the value of a comparative perspective.Mob DNA. 2020 Apr 21;11:16. doi: 10.1186/s13100-020-00215-x. eCollection 2020. Mob DNA. 2020. PMID: 32336995 Free PMC article. Review.
References
-
- Ciuffi A, Bushman FD. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet. 2006;22:388–395. - PubMed
-
- Bushman FD. Targeting survival: Integration site selection by retroviruses and LTR-retrotransposons. Cell. 2003;115:135–138. - PubMed
-
- Cherepanov P, et al. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–381. - PubMed
-
- Ciuffi A, et al. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11:1287–1289. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

