Conformational-dependent and independent RNA binding to the fragile x mental retardation protein
- PMID: 21772992
- PMCID: PMC3136132
- DOI: 10.4061/2011/246127
Conformational-dependent and independent RNA binding to the fragile x mental retardation protein
Abstract
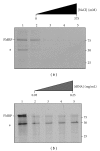
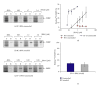
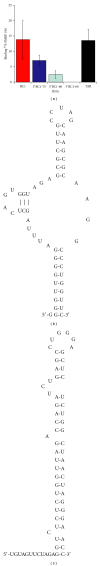
The interaction between the fragile X mental retardation protein (FMRP) and BC1 RNA has been the subject of controversy. We probed the parameters of RNA binding to FMRP in several ways. Nondenaturing agarose gel analysis showed that BC1 RNA transcripts produced by in vitro transcription contain a population of conformers, which can be modulated by preannealing. Accordingly, FMRP differentially binds to the annealed and unannealed conformer populations. Using partial RNase digestion, we demonstrate that annealed BC1 RNA contains a unique conformer that FMRP likely binds. We further demonstrate that this interaction is 100-fold weaker than that the binding of eEF-1A mRNA and FMRP, and that preannealing is not a general requirement for FMRP's interaction with RNA. In addition, binding does not require the N-terminal 204 amino acids of FMRP, methylated arginine residues and can be recapitulated by both fragile X paralogs. Altogether, our data continue to support a model in which BC1 RNA functions independently of FMRP.
Figures


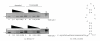
Similar articles
-
On BC1 RNA and the fragile X mental retardation protein.Proc Natl Acad Sci U S A. 2008 Jan 15;105(2):734-9. doi: 10.1073/pnas.0710991105. Epub 2008 Jan 9. Proc Natl Acad Sci U S A. 2008. PMID: 18184799 Free PMC article.
-
Fragile X mental retardation protein (FMRP) binds specifically to the brain cytoplasmic RNAs BC1/BC200 via a novel RNA-binding motif.J Biol Chem. 2005 Sep 30;280(39):33403-10. doi: 10.1074/jbc.M504286200. Epub 2005 Jul 8. J Biol Chem. 2005. PMID: 16006558
-
BC1-FMRP interaction is modulated by 2'-O-methylation: RNA-binding activity of the tudor domain and translational regulation at synapses.Nucleic Acids Res. 2012 May;40(9):4086-96. doi: 10.1093/nar/gkr1254. Epub 2012 Jan 11. Nucleic Acids Res. 2012. PMID: 22238374 Free PMC article.
-
An "Omic" Overview of Fragile X Syndrome.Biology (Basel). 2021 May 13;10(5):433. doi: 10.3390/biology10050433. Biology (Basel). 2021. PMID: 34068266 Free PMC article. Review.
-
RNA Secondary Structure Modulates FMRP's Bi-Functional Role in the MicroRNA Pathway.Int J Mol Sci. 2016 Jun 22;17(6):985. doi: 10.3390/ijms17060985. Int J Mol Sci. 2016. PMID: 27338369 Free PMC article. Review.
Cited by
-
Brain Long Noncoding RNAs: Multitask Regulators of Neuronal Differentiation and Function.Molecules. 2021 Jun 28;26(13):3951. doi: 10.3390/molecules26133951. Molecules. 2021. PMID: 34203457 Free PMC article. Review.
-
Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain.Hum Mol Genet. 2015 Mar 15;24(6):1733-40. doi: 10.1093/hmg/ddu586. Epub 2014 Nov 20. Hum Mol Genet. 2015. PMID: 25416280 Free PMC article.
-
Inherent conformational plasticity in dsRBDs enables interaction with topologically distinct RNAs.Biophys J. 2022 Mar 15;121(6):1038-1055. doi: 10.1016/j.bpj.2022.02.005. Epub 2022 Feb 5. Biophys J. 2022. PMID: 35134335 Free PMC article.
References
-
- Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nature Reviews Neuroscience. 2005;6(5):376–387. - PubMed
-
- Bardoni B, Davidovic L, Bensaid M, Khandjian EW. The fragile X syndrome: exploring its molecular basis and seeking a treatment. Expert Reviews in Molecular Medicine. 2006;8(8):1–16. - PubMed
-
- Jin P, Warren ST. New insights into fragile X syndrome: from molecules to neurobehaviors. Trends in Biochemical Sciences. 2003;28(3):152–158. - PubMed
-
- O’Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annual Review of Neuroscience. 2002;25:315–338. - PubMed
-
- Mercaldo V, Descalzi G, Zhuo M. Fragile X mental retardation protein in learning-related synaptic plasticity. Molecules and Cells. 2009;28(6):501–507. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

