A spectrophotometric assay for conjugation of ubiquitin and ubiquitin-like proteins
- PMID: 21771579
- PMCID: PMC3178097
- DOI: 10.1016/j.ab.2011.06.034
A spectrophotometric assay for conjugation of ubiquitin and ubiquitin-like proteins
Abstract
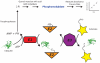
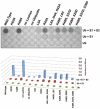
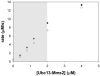
Ubiquitination is a widely studied regulatory modification involved in protein degradation, DNA damage repair, and the immune response. Ubiquitin is conjugated to a substrate lysine in an enzymatic cascade involving an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme, and an E3 ubiquitin ligase. Assays for ubiquitin conjugation include electrophoretic mobility shift assays and detection of epitope-tagged or radiolabeled ubiquitin, which are difficult to quantitate accurately and are not amenable to high-throughput screening. We have developed a colorimetric assay that quantifies ubiquitin conjugation by monitoring pyrophosphate released in the first enzymatic step in ubiquitin transfer, the ATP-dependent charging of the E1 enzyme. The assay is rapid, does not rely on radioactive labeling, and requires only a spectrophotometer for detection of pyrophosphate formation. We show that pyrophosphate production by E1 is dependent on ubiquitin transfer and describe how to optimize assay conditions to measure E1, E2, and E3 activity. The kinetics of polyubiquitin chain formation by Ubc13-Mms2 measured by this assay are similar to those determined by gel-based assays, indicating that the data produced by this method are comparable to methods that measure ubiquitin transfer directly. This assay is adaptable to high-throughput screening of ubiquitin and ubiquitin-like conjugating enzymes.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Robust high-throughput assays to assess discrete steps in ubiquitination and related cascades.BMC Mol Cell Biol. 2020 Mar 30;21(1):21. doi: 10.1186/s12860-020-00262-5. BMC Mol Cell Biol. 2020. PMID: 32228444 Free PMC article.
-
A bioluminescent assay for monitoring conjugation of ubiquitin and ubiquitin-like proteins.Anal Biochem. 2016 Oct 1;510:41-51. doi: 10.1016/j.ab.2016.06.016. Epub 2016 Jun 17. Anal Biochem. 2016. PMID: 27325501
-
Who with whom: functional coordination of E2 enzymes by RING E3 ligases during poly-ubiquitylation.EMBO J. 2020 Nov 16;39(22):e104863. doi: 10.15252/embj.2020104863. Epub 2020 Oct 5. EMBO J. 2020. PMID: 33015833 Free PMC article.
-
Orchestra for assembly and fate of polyubiquitin chains.Essays Biochem. 2005;41:1-14. doi: 10.1042/EB0410001. Essays Biochem. 2005. PMID: 16250894 Review.
-
New insights into ubiquitin E3 ligase mechanism.Nat Struct Mol Biol. 2014 Apr;21(4):301-7. doi: 10.1038/nsmb.2780. Nat Struct Mol Biol. 2014. PMID: 24699078 Review.
Cited by
-
Observing a late folding intermediate of Ubiquitin at atomic resolution by NMR.Protein Sci. 2016 Aug;25(8):1438-50. doi: 10.1002/pro.2940. Epub 2016 May 18. Protein Sci. 2016. PMID: 27111887 Free PMC article.
-
Structural and functional consequences of NEDD8 phosphorylation.Nat Commun. 2021 Oct 12;12(1):5939. doi: 10.1038/s41467-021-26189-9. Nat Commun. 2021. PMID: 34642328 Free PMC article.
-
Mechanisms of Ubiquitin-Nucleosome Recognition and Regulation of 53BP1 Chromatin Recruitment by RNF168/169 and RAD18.Mol Cell. 2017 May 18;66(4):473-487.e9. doi: 10.1016/j.molcel.2017.04.009. Epub 2017 May 11. Mol Cell. 2017. PMID: 28506460 Free PMC article.
-
Chemical proteomic profiling reveals protein interactors of the alarmones diadenosine triphosphate and tetraphosphate.Nat Commun. 2021 Oct 4;12(1):5808. doi: 10.1038/s41467-021-26075-4. Nat Commun. 2021. PMID: 34608152 Free PMC article.
-
Structural basis for GSDMB pore formation and its targeting by IpaH7.8.Nature. 2023 Apr;616(7957):590-597. doi: 10.1038/s41586-023-05832-z. Epub 2023 Mar 29. Nature. 2023. PMID: 36991122 Free PMC article.
References
-
- Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. - PubMed
-
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. - PubMed
-
- Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–86. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

