HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore
- PMID: 21768289
- PMCID: PMC3144403
- DOI: 10.1083/jcb.201012017
HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore
Abstract
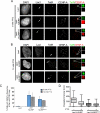
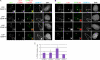
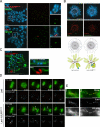
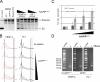
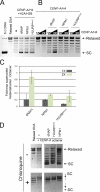
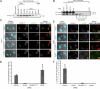
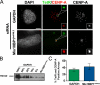
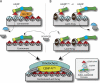
Centromeres of higher eukaryotes are epigenetically marked by the centromere-specific CENP-A nucleosome. New CENP-A recruitment requires the CENP-A histone chaperone HJURP. In this paper, we show that a LacI (Lac repressor) fusion of HJURP drove the stable recruitment of CENP-A to a LacO (Lac operon) array at a noncentromeric locus. Ectopically targeted CENP-A chromatin at the LacO array was sufficient to direct the assembly of a functional centromere as indicated by the recruitment of the constitutive centromere-associated network proteins, the microtubule-binding protein NDC80, and the formation of stable kinetochore-microtubule attachments. An amino-terminal fragment of HJURP was able to assemble CENP-A nucleosomes in vitro, demonstrating that HJURP is a chromatin assembly factor. Furthermore, HJURP recruitment to endogenous centromeres required the Mis18 complex. Together, these data suggest that the role of the Mis18 complex in CENP-A deposition is to recruit HJURP and that the CENP-A nucleosome assembly activity of HJURP is responsible for centromeric chromatin assembly to maintain the epigenetic mark.
Figures
Similar articles
-
Dimerization of the CENP-A assembly factor HJURP is required for centromeric nucleosome deposition.EMBO J. 2013 Jul 31;32(15):2113-24. doi: 10.1038/emboj.2013.142. Epub 2013 Jun 14. EMBO J. 2013. PMID: 23771058 Free PMC article.
-
HJURP interaction with the condensin II complex during G1 promotes CENP-A deposition.Mol Biol Cell. 2017 Jan 1;28(1):54-64. doi: 10.1091/mbc.E15-12-0843. Epub 2016 Nov 2. Mol Biol Cell. 2017. PMID: 27807043 Free PMC article.
-
CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly.J Cell Biol. 2011 Sep 19;194(6):855-71. doi: 10.1083/jcb.201106079. Epub 2011 Sep 12. J Cell Biol. 2011. PMID: 21911481 Free PMC article.
-
Putting CENP-A in its place.Cell Mol Life Sci. 2013 Feb;70(3):387-406. doi: 10.1007/s00018-012-1048-8. Epub 2012 Jun 23. Cell Mol Life Sci. 2013. PMID: 22729156 Free PMC article. Review.
-
Orchestrating the Specific Assembly of Centromeric Nucleosomes.Prog Mol Subcell Biol. 2017;56:165-192. doi: 10.1007/978-3-319-58592-5_7. Prog Mol Subcell Biol. 2017. PMID: 28840237 Free PMC article. Review.
Cited by
-
The cysteine-rich domain in CENP-A chaperone Scm3HJURP ensures centromere targeting and kinetochore integrity.Nucleic Acids Res. 2024 Feb 28;52(4):1688-1701. doi: 10.1093/nar/gkad1182. Nucleic Acids Res. 2024. PMID: 38084929 Free PMC article.
-
The CENP-T C-terminus is exclusively proximal to H3.1 and not to H3.2 or H3.3.Int J Mol Sci. 2015 Mar 12;16(3):5839-63. doi: 10.3390/ijms16035839. Int J Mol Sci. 2015. PMID: 25775162 Free PMC article.
-
Establishment of Centromeric Chromatin by the CENP-A Assembly Factor CAL1 Requires FACT-Mediated Transcription.Dev Cell. 2015 Jul 6;34(1):73-84. doi: 10.1016/j.devcel.2015.05.012. Dev Cell. 2015. PMID: 26151904 Free PMC article.
-
A role for the CAL1-partner Modulo in centromere integrity and accurate chromosome segregation in Drosophila.PLoS One. 2012;7(9):e45094. doi: 10.1371/journal.pone.0045094. Epub 2012 Sep 21. PLoS One. 2012. PMID: 23028777 Free PMC article.
-
Higher-order protein assembly controls kinetochore formation.Nat Cell Biol. 2024 Jan;26(1):45-56. doi: 10.1038/s41556-023-01313-7. Epub 2024 Jan 2. Nat Cell Biol. 2024. PMID: 38168769 Free PMC article.
References
-
- Bergmann J.H., Rodríguez M.G., Martins N.M., Kimura H., Kelly D.A., Masumoto H., Larionov V., Jansen L.E., Earnshaw W.C. 2011. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 30:328–340 10.1038/emboj.2010.329 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

