Protein stability and folding kinetics in the nucleus and endoplasmic reticulum of eucaryotic cells
- PMID: 21767495
- PMCID: PMC3136782
- DOI: 10.1016/j.bpj.2011.05.071
Protein stability and folding kinetics in the nucleus and endoplasmic reticulum of eucaryotic cells
Abstract
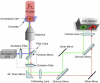
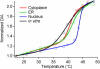
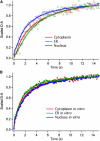
We measure the stability and folding relaxation rate of phosphoglycerate kinase (PGK) Förster resonance energy transfer (FRET) constructs localized in the nucleus or in the endoplasmic reticulum (ER) of eukaryotic cells. PGK has a more compact native state in the cellular compartments than in aqueous solution. Its native FRET signature is similar to that previously observed in a carbohydrate-crowding matrix, consistent with crowding being responsible for the compact native state of PGK in the cell. PGK folds through multiple states in vitro, but its folding kinetics is more two-state-like in the ER, so the folding mechanism can be modified by intracellular compartments. The nucleus increases PGK stability and folding rate over the cytoplasm and ER, even though the density of crowders in the nucleus is no greater than in the ER or cytoplasm. Nuclear folding kinetics (and to a lesser extent, thermodynamics) vary less from cell to cell than in the cytoplasm or ER, indicating a more homogeneous crowding and chemical environment in the nucleus.
Copyright © 2011 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
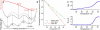

Comment in
-
Protein folding inside the cell.Biophys J. 2011 Jul 20;101(2):265-6. doi: 10.1016/j.bpj.2011.06.018. Biophys J. 2011. PMID: 21767477 Free PMC article. No abstract available.
Similar articles
-
Protein folding inside the cell.Biophys J. 2011 Jul 20;101(2):265-6. doi: 10.1016/j.bpj.2011.06.018. Biophys J. 2011. PMID: 21767477 Free PMC article. No abstract available.
-
Subcellular modulation of protein VlsE stability and folding kinetics.FEBS Lett. 2016 May;590(10):1409-16. doi: 10.1002/1873-3468.12193. Epub 2016 May 17. FEBS Lett. 2016. PMID: 27129718
-
The extracellular protein VlsE is destabilized inside cells.J Mol Biol. 2014 Jan 9;426(1):11-20. doi: 10.1016/j.jmb.2013.08.024. Epub 2013 Sep 4. J Mol Biol. 2014. PMID: 24013077 Free PMC article.
-
Denatured states of yeast phosphoglycerate kinase.Biochemistry (Mosc). 1998 Mar;63(3):259-75. Biochemistry (Mosc). 1998. PMID: 9526123 Review.
-
The many functions of the endoplasmic reticulum chaperones and folding enzymes.IUBMB Life. 2014 May;66(5):318-26. doi: 10.1002/iub.1272. Epub 2014 May 19. IUBMB Life. 2014. PMID: 24839203 Review.
Cited by
-
Excluded-volume effects in living cells.Angew Chem Int Ed Engl. 2015 Feb 16;54(8):2548-51. doi: 10.1002/anie.201409847. Epub 2014 Dec 29. Angew Chem Int Ed Engl. 2015. PMID: 25557778 Free PMC article.
-
Exploring the dynamics and structure of PpiB in living Escherichia coli cells using electron paramagnetic resonance spectroscopy.Protein Sci. 2024 Mar;33(3):e4903. doi: 10.1002/pro.4903. Protein Sci. 2024. PMID: 38358137 Free PMC article.
-
Chemical interactions modulate λ6-85 stability in cells.Protein Sci. 2023 Jul;32(7):e4698. doi: 10.1002/pro.4698. Protein Sci. 2023. PMID: 37313657 Free PMC article.
-
Quantifying the thermodynamics of protein unfolding using 2D NMR spectroscopy.Commun Chem. 2020 Aug 7;3:100. doi: 10.1038/s42004-020-00358-1. Commun Chem. 2020. PMID: 33718626 Free PMC article.
-
In silico protein dynamics in the human cytoplasm: Partial folding, misfolding, fold switching, and non-native interactions.Protein Sci. 2023 Nov;32(11):e4790. doi: 10.1002/pro.4790. Protein Sci. 2023. PMID: 37774143 Free PMC article.
References
-
- Kubelka J., Hofrichter J., Eaton W.A. The protein folding ‘speed limit’. Curr. Opin. Struct. Biol. 2004;14:76–88. - PubMed
-
- Lange O.F., Lakomek N.A., de Groot B.L. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471–1475. - PubMed
-
- Henzler-Wildman K.A., Lei M., Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450:913–916. - PubMed
-
- Bryngelson J.D., Onuchic J.N., Wolynes P.G. Funnels, pathways, and the energy landscape of protein folding: a synthesis. Proteins. 1995;21:167–195. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

